Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LUO Danli(罗丹丽), LIU Yuan(刘媛), HUI Min(惠敏), SONG Chengwen(宋呈文), LIU Hourong(刘厚荣), CUI Zhaoxia(崔朝霞)
- Molecular characterization and expression profiles of four transformer-2 isoforms in the Chinese mitten crab Eriocheir sinensis
- Chinese Journal of Oceanology and Limnology, 35(4): 782-791
- http://dx.doi.org/10.1007/s00343-017-6071-z
Article History
- Received Mar. 12, 2016
- accepted in principle May. 3, 2016
- accepted for publication Jun. 20, 2016
2 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266071, China;
3 National & Local Joint Engineering Laboratory of Ecological Mariculture, Qingdao 266071, China;
4 University of Chinese Academy of Sciences, Beijing 100049, China
The transformer-2 (tra-2) gene encodes a splicing regulator that can be involved in the sex determination and differentiation pathway of Drosophila melanogaster in a tissue-specific manner (Mattox et al., 1990). In the female soma, tra-2 protein acts with transformer (tra) to direct pre-mRNA splicing of the doublesex (dsx) gene in the somatic sex-determination cascade (Verhulst et al., 2010) and sets downstream regulators into the female mode (Schütt and Nothiger, 2000; Erickson and Quintero, 2007). In the male germline, tra-2 autoregulates the self-splicing of germline-specific tra-2 transcripts, and performs an essential function during spermatogenesis (McGuffin et al., 1998; Chandler et al., 2001).
The tra-2 gene has been studied in many insects, including Drosophila virilis (Chandler et al., 1997), Musca domestica (Burghardt et al., 2005), Bombyx mori (Niu et al., 2005), Ceratitis capitata (Gomulski et al., 2008), Anastrepha species (Sarno et al., 2010) and Tribolium castaneum (Shukla and Palli, 2013). The tra-2 gene has also been identified in mammals (human) and birds (chicken) as an mRNA splicing regulator (Dauwalder et al., 1996; Yamamoto et al., 2002). In crustacean species, a tra homolog was cloned in the giant tiger shrimp Penaeus monodon (Li et al., 2012) and the oriental river prawn Macrobrachium nipponense (Zhang et al., 2013), and no sexual dimorphism of the alternative splicing variants and expression patterns was observed. However, one of three tra-2 isoforms in the Chinese shrimp Fenneropenaeus chinensis exhibited a particularly high expression level in the ovary and it was speculated to participate in sex determination (Li et al. 2012). Previously, no tra-2 gene has been identified in crabs.
The Chinese mitten crab Eriocheir sinensis is one of the most important aquaculture crustaceans in China. Like many other crustaceans, E. sinensis shows sexually dimorphic traits. For example, male crabs are usually larger. In the present study, four tra-2 isoforms, designated as Estra-2a, Estra-2b, Estra-2c and Estra-2d, were isolated by screening the embryonic and juvenile transcriptomes of E. sinensis (Liu et al., 2015). We characterize the nucleotide and amino acid sequences of the four tra-2 genes and examine their expression profiles in early developmental stages and adult tissues of E. sinensis.
2 MATERIAL AND METHOD 2.1 Animal and sample preparationMitten crabs were obtained from a farm in Panjin, China. Samples of embryonic developmental stages were collected from the pleopods of berried females. Eggs were examined under a microscope to identify embryonic developmental stages (Du et al., 1992), including fertilized egg stage, 2–4 cell stage, blastula stage, gastrula stage and egg-nauplius stage. The larvae were sampled from zoea Ⅰ, zoea Ⅱ, zoea Ⅲ, zoea Ⅳ, zoea Ⅴ and megalopa stages. Crab tissues, including muscle, hemocytes, hepatopancreas, gill, muscle, heart, thoracic ganglia, testis and ovary were sampled from eight healthy adult crabs (four males and four females), and then dissected and preserved in liquid nitrogen for RNA or DNA extraction.
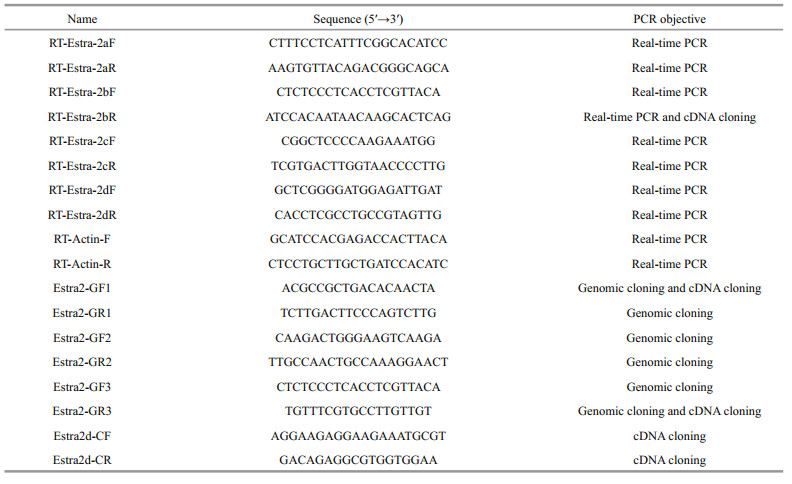
2.2 RNA extraction, cDNA synthesis and sequence determination of Estra-2Total RNA was isolated from each of the embryonic and larval developmental stages and adult tissues with Trizol reagent (Invitrogen, USA), and then treated with RQ1 RNase-Free DNase I (Promega, USA) to remove DNA contamination. RNA quality and concentration were determined by spectrophotometry and electrophoresis (1% agarose gel). The first-strand complementary DNAs (cDNAs) were reverse transcribed from 2-μg RNA samples by Moloney-Murine Leukemia Virus (M-MLV) reverse transcriptase (Promega) according to the manufacturer's protocols. The cDNA sequence of Estra-2 isoforms were amplified using the cDNA template with four pairs of primers: Estra2-GF1 and Estra2-GR1; Estra2-GF1 and RT-Estra-2bR; Estra2-GF1 and Estra2-GR3; and Estra2d-CF and Estra2-CR. PCR reaction mixtures (25 μL) contained 19.8 μL sterile distilled H2O, 2.5 μL of 10× PCR buffer, 0.5 μL of dNTP (10 mol/L), 0.5 μL of each primer (10 mol/L), 0.2 mL (1 U) of Taq polymerase (TaKaRa, Japan), and 1 μL of DNA template (approximately 30 ng). PCR amplification was performed using the following conditions: an initial denaturation at 94℃ for 3 min 30 s, followed by 34 cycles of denaturation at 94℃ for 30 s, annealing at 55℃ for 50 s, extension at 72℃ for 1 min, and a final extension at 72℃ for 10 min. The PCR products were gel-purified and sequenced.
2.3 DNA extraction and genomic DNA amplification of Estra-2a, Estra-2b and Estra-2cGenomic DNA was extracted from muscle using the phenol/chloroform method (Sambrook and Russell, 2001). The genomic sequences of Estra-2a, Estra-2b and Estra-2c were amplified using the DNA template with three pairs of primers: Estra2-GF1 and Estra2-GR1; Estra2-GF2 and Estra2-GR2; and Estra2-GF3 and Estra2-GR3 (Table 1). PCR reaction mixtures (25 μL) contained 19.8 μL sterile distilled H2O, 2.5 μL of 10× PCR buffer, 0.5 μL of dNTP (10 mol/L), 0.5 μL of each primer (10 mol/L), 0.2 mL (1 U) of Taq polymerase (TaKaRa), and 1 μL of DNA template (approximately 30 ng). The PCR amplification was performed using the following conditions: an initial denaturation at 94℃ for 3 min 30 s, followed by 34 cycles of denaturation at 94℃ for 30 s, annealing at 55℃ for 50 s, extension at 72℃ for 2 min 30 s, and a final extension at 72℃ for 10 min. The PCR products were gel-purified and sequenced. The obtained sequences were assembled by the software SeqMan 5.01.
Using the BlastX algorithm, four unigenes from the embryonic and juvenile transcriptomes of E. sinensis were found to be highly homologous to the tra-2 genes of the shrimp F. chinensis and P. monodon. The deduced amino acid sequence and open reading frames (ORF) were analyzed with the Expert Protein Analysis System (http://www.expasy.org/). The molecular weights and isoelectric points of deduced amino acid sequences were determined using the Compute pI/Mw tool (http://web.expasy.org/ compute_pi/). Genomic organizations were performed using the Genewise tool (http://www.ebi.ac.uk/ Wise2/index.html). Sequence alignment was carried out with online ClustalW2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Neighbor-joining tree with bootstrap values were constructed for phylogenetic analysis using MEGA software version 4.0 (Tamura et al., 2007).
2.5 Expression of Estra-2 isoforms in different developmental stages and adult tissuesFour pairs of gene-specific primers were designed for expression analysis of four isoforms of tra-2: RTEstra-2aF and RT-Estra-2aR (278 bp amplicons); RTEstra-2bF and RT-Estra-2bR (262 bp amplicons); RTEstra-2cF and RT-Estra-2cR (314 bp amplicons); and RT-Estra-2dF and RT-Estra-2dR (126 bp amplicons) (Table 1). The real-time PCR product of each isoform was cloned and sequenced. Quantitative real-time PCR was carried out using the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA). Reactions were performed in triplicate for each sample and the expression levels were normalized to that of β-actin from E. sinensis (GenBank accession no. HM053699). PCR was carried out using the following conditions: 95℃ for 30 s, followed by 40 cycles of 95℃ for 5 s and 60℃ for 31 s. PCR reaction mixtures (25 μL) contained 5 μL of 2× SYBR Premix Ex TaqTM (TaKaRa), 0.2 μL 50× ROX Reference Dye Ⅱ, 4 μL of the diluted cDNA, 0.2 μL of each primer (10 μmol/L), and 0.4 μL of RNase-free H2O. The threshold cycle value between 15 and 30 was used for further analysis.
2.6 Statistical analysisFold changes of gene expression relative to controls were analyzed using the 2-ΔΔCt method (Livak and Schmittgen, 2001). Data are presented in terms of relative mRNA expression as means±S.E. The results were subjected to one-way analysis of variance (oneway ANOVA) followed by a post hoc Tukey's multiple-group comparison test or Student's t test using SPSS 16.0; P < 0.05 or < 0.01 were considered statistically significant.
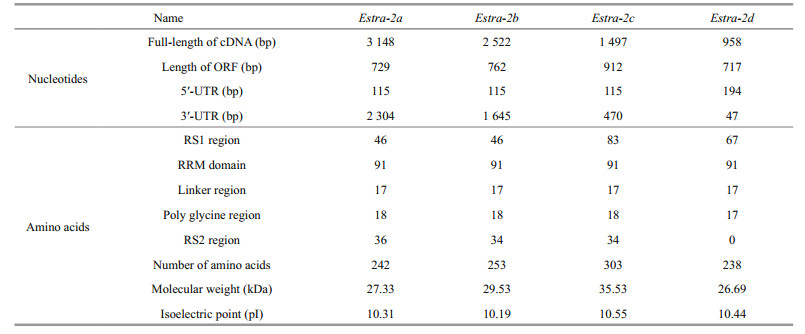
3 RESULT 3.1 Sequence organization of Estra-2 isoformsThe full-length cDNA sequences of Estra-2a, Estra-2b and Estra-2c were 3 148, 2 522 and 1 497 bp, and were deposited in GenBank under accession numbers KU291992, KU291993 and KU291994, respectively. They all shared the same 5′-UTR with a length of 115 bp. Estra-2a contained 2 304 bp of 3′-UTR and 729 bp of open-reading frame (ORF) corresponding to 242 amino acids. Estra-2b included 1 645 bp of 3′-UTR and 762 bp of ORF corresponding to 253 amino acids, while Estra-2c comprised 470 bp of 3′-UTR and 912 bp of ORF corresponding to 303 amino acids. Two RS domains (RS1 and RS2), a conserved RNA-recognition motif (RRM) domain (91 amino acids) and a linker region (17 amino acids) typically were present in Estra-2a, Estra-2b and Estra-2c.
The full-length cDNA sequence of another tra-2 homolog, Estra-2d, was 958 bp in length with the 5′-UTR of 194 bp, 3′-UTR of 47 bp and an ORF of 717 bp, corresponding to 238 amino acids (GenBank accession No. KU291995). Estra-2d exhibited low similarity to the other three isoforms in cDNA sequence, but high identity in amino acid sequence of the conserved domain. The conserved RRM domain (91 amino acids) and linker region (17 amino acids) were also found in Estra-2d, but only one RS region (RS1) with 67 amino acids (Pro22–Ser81) was identified. Detailed characteristics of the isolated cDNAs and the corresponding putative proteins are summarized in Table 2.
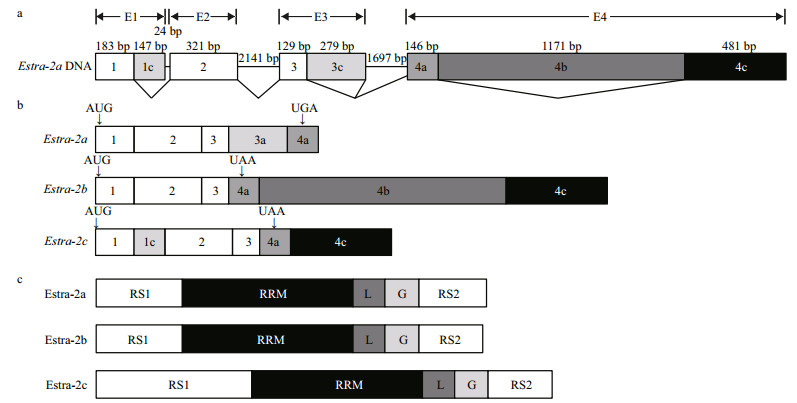
The genetic structures of Estra-2a Estra-2b and Estra-2c indicate that they originated from the same genomic DNA sequence through alternative splicing (Fig. 1). Three PCR products were obtained and sequenced (514 bp, 2 665 bp and 3 764 bp, respectively). Assembly of the DNA sequences of the PCR products produced a genomic DNA fragment of Estra-2a, Estra-2b and Estra-2c (a total of 6 719 bp, GenBank accession No. KU257540). As shown in Fig. 1, the genomic DNA sequence contained four exons (E1, E2, E3 and E4) and three introns. The first exon (E1) was divided into two parts, exon1 and exon1c. All three mature mRNAs contained exon1, while Estra-2c retained a unique exon1c. The second exon (E2) was identical in all mature mRNAs. The third exon (E3) consisted of two parts (exon3 and exon3a), with exon3 conserved in all three variants and exon3a present only in Estra-2a. The forth exon (E4) comprised three parts (exon4a, exon4b and exon4c). Only exon4a was retained in mature Estra-2a mRNA. Estra-2b contained the full length of E4. The exon4b was absent from Estra-2c (Fig. 1b).
|
| Figure 1 Schematic presentation of the DNA, mature mRNA and deduced amino acid sequences of Estra-2 isoforms from E.sinensis a. genomic DNA sequence organization of Estra-2a, Estra-2b and Estra-2c. The boxes with numbers show the exons and the lines show the introns. The length of each exon or intron is displayed above the box or line, respectively. Alternative splicings of Estra-2a, Estra-2b and Estra-2c pre-mRNA are shown by lines connecting the boxes; b. the full-length mature mRNAs of Estra-2a, Estra-2b and Estra-2c. Exons are numbered corresponding to (a). The start codon (AUG) and stop codons (UGA and UAA) are marked with arrows above the exon boxes; c. deduced amino acid sequence of Estra-2a, Estra-2b and Estra-2c with predicted domains. The RRM box indicates the conserved RNA recognition motif, while L, RS1 and RS2 show the linker region and arginine/serine rich regions 1 and 2, respectively. |
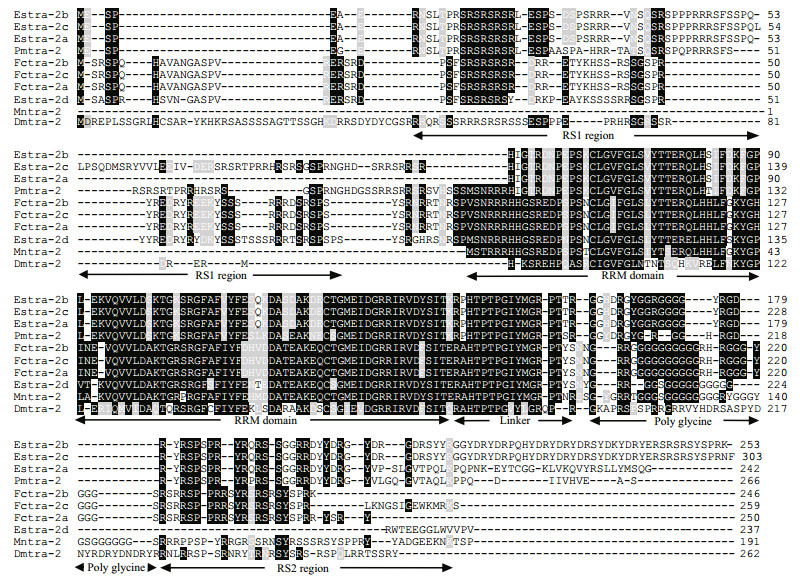
The amino acid sequences corresponding to all three isoforms included two arginine/serine-rich (RS) regions (RS1 and RS2). Estra-2c retained exon3a and produced a longer RS1 region with 83 amino acids (Arg8-Arg90), compared with the shorter RS1 region in Estra-2a and Estra-2b with 46 amino acids (Arg8-Gln89). The RS2 region comprised 25 amino acids (Arg180-Tyr204 in Estra-2a and Estra-2b; Arg229-Tyr253 in Estra-2c) in all three variants.
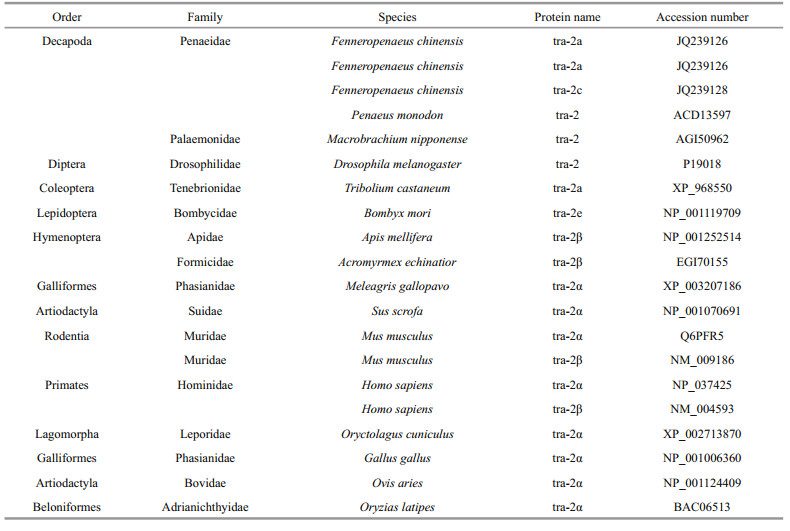
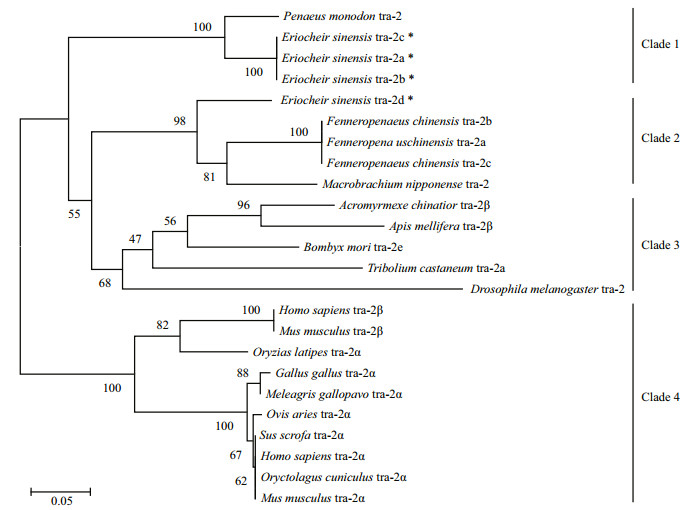
3.3 Multiple sequence alignment and phylogenetic analysisBLAST analysis based on amino acid sequences showed that Estra-2a, Estra-2b and Estra-2c displayed 74%, 72% and 74% identity, respectively, with Pmtra-2 (P. monodon, ACD13597). Estra-2d had 87% identity with Mntra-2 (Macrobrachium nipponense, AGI50962) and 72% identity with Fctra-2 (F. chinensis, AFU60540-2). The RRM domain and linker region shared high identity with tra-2 from other species, while the sequence similarities of the RS1 and RS2 regions with those in other species were very low (Fig. 2). A phylogenetic tree was constructed by the Bayesian method, based on 20 amino acid sequences of tra-2 members (Table 3). Estra-2 isoforms were classified into a single clade with tra-2 sequences from insects and decapods (Fig. 3). The four isoforms were divided into two clades with Estra-2a, Estra-2b and Estra-2c clustered into a sub-clade with the shrimp P. monodon, and Estra-2d grouped with the shrimp F. chinensis and M. nipponense (Fig. 3).
|
| Figure 2 Multiple sequence alignments of Estra-2 isoforms with tra-2 from other species The deduced amino acid sequences are summarized in Table 2. Identical sequences are shown in dark gray and similar sequences in light gray. RS1, RRM domain, linker (L), poly Glycine and RS2 regions are defined according to SMART analysis. |
|
| Figure 3 Neighbor-joining phylogenetic tree of tra-2 based on amino acid sequences The sequences used in the phylogenetic tree are summarized in Table 2. Numbers in the branches represent the bootstrap values (%) from 1000 replicates. Three Estra-2s of E. sinensis are marked with asterisks. |
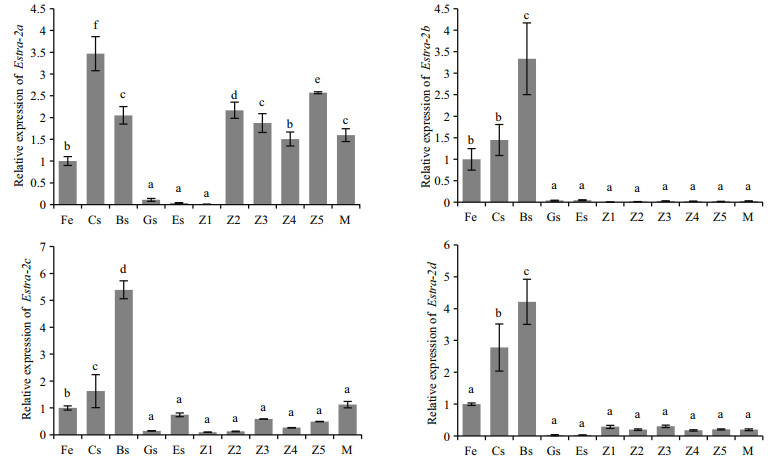
Temporal expression patterns of Estra-2 mRNA in embryonic and larval stages of the crab were detected by qRT-PCR. The four Estra-2 isoforms exhibited similar expression patterns from the fertilized egg stage to the egg-nauplius stage (Fig. 4). They were all detected in fertilized eggs, up-regulated in the cleavage and blastula stages (P≤0.01), and markedly downregulated at the gastrula stage (P≤0.01), remaining steady until the egg-nauplius stage. During the larval stages, the expression of Estra-2a was sharply elevated at the zoea Ⅱ stage (P≤0.01), and then fluctuated at a high level until the megalopa stage (P≤0.01). Estra-2b and Estra-2d stabilized at a low expression level, while Estra-2c gradually increased throughout the whole larval stage from zoea Ⅰ to megalopa.
|
| Figure 4 Temporal expression of Estra-2 isoforms in the embryonic and larval stages of E. sinensis Fe: fertilized egg stage; Cs: 2–4 cell stage; Bs: blastula stage; Gs: gastrula stage; Es: egg-nauplius stage; Z1: zoea Ⅰ; Z2: zoea Ⅱ; Z3: zoea Ⅲ; Z4: zoea Ⅳ; Z5: zoea Ⅴ; M: megalopa. Vertical bars represent the mean±S.E. (n=3). Significant differences among stages are indicated by different letter labels (one-way ANOVA followed by post hoc Tukey's multiple group comparison P≤0.05). |
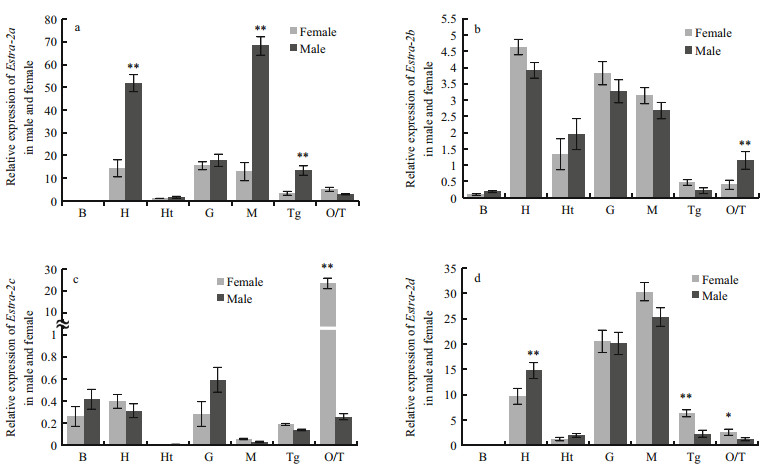
To verify whether Estra-2 isoforms showed sexual dimorphism, qPCR was carried out in various tissues, including hemocytes, hepatopancreas, heart, gill, muscle, thoracic ganglia, testis and ovary, in male and female adults (Fig. 5). Estra-2a, Estra-2b and Estra-2d were all highly expressed in somatic tissues. However, Estra-2a displayed significantly higher expression in several tissues of males than in females, particularly hepatopancreas, muscle and thoracic ganglia (P≤0.01; Fig. 5a). In the gonads, the expression of Estra-2a was relatively low, while Estra-2b showed small sex differences with a higher expression level in the testis than in the ovary (P≤0.01; Fig. 5b and d). Estra-2c exhibited a different expression pattern with its highest expression in the ovary and low expression in somatic tissues (P≤0.01; Fig. 5c).
|
| Figure 5 Tissue distributions of four Estra-2 isoforms in male and female E. sinensis B: hemocytes; H: hepatopancreas; Ht: heart; G: gill; M: muscle; Tg: thoracic ganglia; T: testis; O: ovary. Vertical bars represent means±S.E. (n=3). Significant differences between males and females are indicated with single (P≤0.05) or double (P≤0.01) asterisks (Student's t test). |
The identification of four Estra-2 isoforms from two genomic loci in E. sinensis is the first report of tra-2 genes in a crab. Of these, Estra-2a, Estra-2b and Estra-2c are generated by alternative splicing of the same pre-mRNA. Alternative splicing of tra-2 genes is also found in other arthropod species, including D. melanogaster (Mattox et al., 1990), Bombyx mori (Niu et al., 2005), Sciara ocellaris (Martín et al., 2011), and F. chinensis (Li et al., 2012). However, no splicing variants and a single tra-2 gene were reported in M. nipponense (Zhang et al., 2013) and P. monodon (Leelatanawit et al., 2009). Furthermore, the alternative splicing pattern of Estra-2 pre-mRNA in the last exon resembles that of Dmtra-2, Bmtra-2 and Fctra-2 pre-mRNAs.
Further analysis of the predicted amino acid sequence alignments shows that the diversity of Estra-2 isoforms is mainly observed in the RS domains. Estra-2c contained a longer RS1 region than the other three isoforms, while the RS2 region was absent from Estra-2d. The RS domain mediates splice-site recognition and splicing regulation (Long and Caceres, 2009) and its potency is proportional to the number of RS-dipeptide repeats (Philipps et al., 2003). The numbers of RS-dipeptide repeats in RS1 reported in other decapods are: one in M. nipponense (Zhang et al., 2013), eight in F. chinensis (Li et al., 2012) and 13 in P. monodon (Leelatanawit et al., 2009). Estra-2c produced the largest RS1 region within 12 RSdipeptide repeats, compared with only six RSdipeptide repeats in the other three isoforms (Fig. 1c). As suggested by Philipps et al. (2003), we speculate that Estra-2c might have stronger splicing activation potency than the other three isoforms. Considering the role of the RS2 region in D. melanogaster (Blencowe et al., 1999) the lack of RS2 in Estra-2d suggests that Estra-2d has no function in pre-mRNA splicing. The RRM domain is indispensable for splicing, mediates RNA binding and determines substrate specificity for individual SR-related proteins (Long and Caceres, 2009). The existence of the RRM domain in Estra-2 isoforms might provide a structural basis for their potential function as a splicing factor.
The temporal expression analysis revealed that Estra-2 transcripts are present throughout the early developmental stages of E. sinensis, especially in early embryonic stages. These stages are mainly controlled by maternal products before the zygotic genome is activated (Schier, 2007). Therefore, we infer that the Estra-2 transcripts are maternally derived and might help to maintain the accumulation of tra in the embryo germline, as maternal tra-2s do in insects (McGuffin et al., 1998; Chandler et al., 2001; Burghardt et al., 2005).
Estra-2a in our study is the first tra-2 gene reported to show differential expression between the sexes in somatic tissues. However, in contrast to the function of Dmtra-2s in the female soma of D. melanogaster (Mattox et al., 1990), Estra-2a was highly accumulated in male somatic tissues and might be involved in the sexual differentiation of males. In contrast to the other three isoforms, Estra-2c transcripts were most abundant in the ovary, which is consistent with reports on other crustaceans, e.g., shrimp F. chinensis (Li et al., 2012). Crustaceans are a sister group of insects (Boore et al., 1998). Considering the function of tra-2 in insects, including D. melanogaster (Mattox et al., 1990), M. domestica (Burghardt et al., 2005) and Apis mellifera (Gempe et al., 2009), these expression patterns suggest that Estra-2c functions in gonadal development in the mitten crab. The activity of tra-2 was considered to be regulated through sex-specific interaction with tra protein rather than through the presence or absence of sex-specific variants in D. melanogaster (Mattox and Baker, 1991). However, the ubiquitous but differential distribution of Estra-2 transcripts in E. sinensis provides a possibility that tra-2 may have multifunctional roles in different tissues of this economically important species.
5 CONCLUSIONIn summary, we obtained four tra-2 isoforms from the transcriptomes of E. sinensis, designated as: Estra-2a, Estra-2b, Estra-2c and Estra-2d. Estra-2a and Estra-2c are suggested to be involved in sexual differentiation in the male soma and ovary. The identification of tra-2 isoforms in E. sinensis lays a foundation for further study of the sex-determining mechanisms of crabs.
6 ACKNOWLEDGEMENTWe were grateful to all laboratory members for technical advice and helpful discussions.
| Blencowe B J, Bowman J A L, McCracken S, Rosonina E, 1999. SR-related proteins and the processing of messenger RNA precursors. Biochemistry and Cell Biology, 77(4): 277–291. Doi: 10.1139/o99-048 |
| Boore J L, Lavrov D V, BrownW M, 1998. Gene translocation links insects and crustaceans. Nature, 392(6677): 667–668. Doi: 10.1038/33577 |
| Burghardt G, Hediger M, Siegenthaler C, Moser M, Dübendorfer A, Bopp D, 2005. The transformer2 gene in Musca domestica is required for selecting and maintaining the female pathway of development. Development Genes and Evolution, 215(4): 165–176. Doi: 10.1007/s00427-004-0464-7 |
| Chandler D S, McGuffin M E, Mattox W, 2001. Functionally antagonistic sequences are required for normal autoregulation of Drosophila tra-2 pre-mRNA splicing. Nucleic Acids Research, 29(14): 3012–3019. Doi: 10.1093/nar/29.14.3012 |
| Chandler D S, McGuffin M E, Piskur J, Yao J, Baker B S, Mattox W, 1997. Evolutionary conservation of regulatory strategies for the sex determination factor transformer-2. Molecular and Cellular Biology, 17(5): 2908–2919. Doi: 10.1128/MCB.17.5.2908 |
| Dauwalder B, Amaya-Manzanares F, Mattox W, 1996. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proceedings of the National Academy of Sciences of the United States of America, 93(17): 9004–9009. Doi: 10.1073/pnas.93.17.9004 |
| Du N S, Zhao Y L, Lai W. 1992. Transactions of the Chinese Crustacean Society. A Study on the Embryonic Development of the Chinese Mitten-Handed Crab, Eriocheir Sinensis (Crustacea: Decapoda). 2st edn. Qidao Ocean University Press, Qingdao, China. p. 128-135. (in Chinese) |
| Erickson J W, Quintero J J, 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:a ratio, signals sex in Drosophila. PLoS Biology, 5(12): e332. Doi: 10.1371/journal.pbio.0050332 |
| Gempe T, Hasselmann M, Schiøtt M, Hause G, Otte M, Beye M, 2009. Sex determination in honeybees:two separate mechanisms induce and maintain the female pathway. PLoS Biology, 7(10): e1000222. Doi: 10.1371/journal.pbio.1000222 |
| Gomulski L M, Dimopoulos G, Xi Z Y, Soares M B, Bonaldo M F, Malacrida A R, Gasperi G. 2008. Gene discovery in an invasive tephritid model pest species, theMediterranean fruit fly, Ceratitis capitata. BMC Genomics, 9: 243, http://dx.doi.org/10.1186/1471-2164-9-243. |
| Leelatanawit R, Sittikankeaw K, Yocawibun P, Klinbunga S, Roytrakul S, Aoki T, Hirono I, Menasveta P. 2009. Identification, characterization and expression of sexrelated genes in testes of the giant tiger shrimp Penaeus monodon. Comparative Biochemistry and Physiology Part A:Molecular & Integrative Physiology, 152(1):66-76. |
| Li S, Li F, Wen R, Xiang J, 2012. Identification and characterization of the sex-determiner transformer-2 homologue in Chinese Shrimp, Fenneropenaeus chinensis. Sexual Development, 6(5): 267–278. Doi: 10.1159/000341377 |
| Liu Y, Hui M, Cui Z X, Luo D L, Song C W, Li Y D, Liu L, 2015. Comparative transcriptome analysis reveals sexbiased gene expression in juvenile Chinese mitten crab Eriocheir sinensis. PLoS One, 10(7): e0 133 068. Doi: 10.1371/journal.pone.0133068 |
| Livak K J, Schmittgen T D, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4): 402–408. Doi: 10.1006/meth.2001.1262 |
| Long J C, Caceres J F, 2009. The SR protein family of splicing factors:master regulators of gene expression. Biochemical Journal, 417(1): 15–27. Doi: 10.1042/BJ20081501 |
| Martín I, Ruiz M F, Sánchez L. 2011. The gene transformer-2 of Sciara (Diptera, Nematocera) and its effect on Drosophila sexual development. BMC Developmental Biology, 11: 19, http://dx.doi.org/10.1186/1471-213X-11-19. |
| Mattox W, Baker B S, 1991. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes & Development, 5(5): 786–796. |
| Mattox W, Palmer M J, Baker B S, 1990. Alternative splicing of the sex determination gene transformer-2 is sexspecific in the germ line but not in the soma. Genes & Development, 4(5): 789–805. |
| McGuffin M E, Chandler D, Somaiya D, Dauwalder B, Mattox W, 1998. Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics, 149(3): 1477–1486. |
| Niu B L, Meng Z Q, Tao Y Z, Lu S L, Weng H B, He L H, Shen W F, 2005. Cloning and alternative splicing analysis of Bombyx mori transformer-2 gene using silkworm EST database. Acta Biochimica et Biophysica Sinica, 37(11): 728–736. Doi: 10.1111/abbs.2005.37.issue-11 |
| Philipps D, Celotto A M, Wang Q Q, Tarng R S, Graveley B R, 2003. Arginine/serine repeats are sufficient to constitute a splicing activation domain. Nucleic Acids Research, 31(22): 6502–6508. Doi: 10.1093/nar/gkg845 |
| Sambrook J, Russell D W. 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York. 545p. |
| Sarno F, Ruiz M F, Eirín-López J M, Perondini A L P, Selivon D, Sánchez L, 2010. The gene transformer-2 of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. BMC Developmental Biology, 10: 140. |
| Schier A F. 2007. The maternal-zygotic transition: death and birth of RNAs. Science, 316(5823): 406-407, http://dx.doi.org/10.1126/science.1140693. |
| Schütt C, Nothiger R, 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development, 127(4): 667–677. |
| Shukla J N, Palli S R, 2013. Tribolium castaneum Transformer-2 regulates sex determination and development in both males and females. Insect Biochemistry and Molecular Biology, 43(12): 1125–1132. Doi: 10.1016/j.ibmb.2013.08.010 |
| Tamura K, Dudley J, Nei M, Kumar S, 2007. MEGA4:molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24(8): 1596–1599. Doi: 10.1093/molbev/msm092 |
| Verhulst E C, Van de Zande L, Beukeboom L W, 2010. Insect sex determination:it all evolves around transformer. Current Opinion in Genetics & Development, 20(4): 376–383. |
| Yamamoto I, Tsukada A, Saito N, Shimada K, 2002. cDNA cloning and mRNA expression of Transformer 2 (Tra 2) in chicken embryo. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression, 1579(2-3): 185–188. Doi: 10.1016/S0167-4781(02)00511-0 |
| Zhang Y P, Fu H T, Qiao H, Jin S B, Jiang S F, Xiong Y W, Gong Y S, Zhang X Z, 2013. Molecular cloning and expression analysis of transformer-2 gene during development in Macrobrachium nipponense (de Haan 1849). Journal of the World Aquaculture Society, 44(3): 338–349. Doi: 10.1111/jwas.2013.44.issue-3 |
 2017, Vol. 35
2017, Vol. 35