Institute of Oceanology, Chinese Academy of Sciences
Article Information
- TANG Xianghai(唐祥海), XU Kuipeng(徐奎鹏), HAN Xiaojuan(韩晓娟), MO Zhaolan(莫照兰), MAO Yunxiang(茅云翔)
- Complete genome of Cobetia marina JCM 21022T and phylogenomic analysis of the family Halomonadaceae
- Chinese Journal of Oceanology and Limnology, 36(2): 528-536
- http://dx.doi.org/10.1007/s00343-017-6239-6
Article History
- Received Sep. 4, 2016
- accepted in principle Sep. 8, 2016
- accepted for publication Sep. 14, 2016
2 Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071, China;
3 CAS Key Laboratory of Biobased Materials, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences, Qingdao 266101, China
Cobetia marina is a gram-negative marine bacterium that is used as a model organism in marine biofouling studies for its typical obligatory aerobic characteristics that allows easy culturing and handling. Further, biofilms formed by C. marina can influence the secondary colonization of invertebrates and algae (Shea et al., 1995). The chronological record of the taxonomy shows that C. marina has hitherto been classified and revised many times. Although all the classifications referred to the same species, it was first identified as Arthrobacter marinus (Cobet et al., 1970), and subsequently described as Pseudomonas marina (Baumann et al., 1972), Delaya marina (Baumann et al., 1983; Shea et al., 1995; Ista et al., 1996) and Halomonas marina (Dobson and Franzmann, 1996; Ista et al., 1999; Mata et al., 2002). More than ten years ago, it was suggested that Halomonas marina should be assigned to a new genus Cobetia in the family Halomonadaceae based on a comparative sequence analysis of 23S and 16S rRNA (Arahal et al., 2002a, b) sequences. This was because the Halomonas marina sequences were too phylogenetically distant from those of other Halomonas species to be considered as belonging to the same genus.
To date, although many molecular studies have been carried out to understand the phylogenetic relationship of Halomonadaceae species, these studies were based mainly on 23S and 16S rDNA sequences (Dobson and Franzmann, 1996; Arahal et al., 2002a, b; Ntougias et al., 2007; de la Haba et al., 2010; Romanenko et al., 2013). Therefore, to better understand the C. marina surface-associated lifestyle and the phylogeny of the family Halomonadaceae, it is important to sequence the whole genomes of representative species and strains. The genome sequences of several Halomonadaceae species have been reported recently (Copeland et al., 2011; Schwibbert et al., 2011; Sánchez-Porro et al., 2013; Sharko et al., 2016), including the genome of C. marina KMM 296 (Balabanova et al., 2016a) that was afterward reclassified as C. amphilecti KMM 296 (Balabanova et al., 2016b). In this study, we used the publicly available genome information to conduct comparative genomics and phylogenomics analyses. On strategy of genome sequencing and assembly, the single molecule real-time sequencing technology (SMRT) was chosen in this study, which is a sequencing-by-synthesis technology based on realtime imaging of fluorescently tagged nucleotides as they are synthesized along individual DNA template molecules. SMRT can produces long and unbiased sequences, and ensure assembly of complex repeat structures and GC and AT rich regions that are often hard to assemble in short-read sequencing technology (Eid et al., 2009; Roberts et al., 2013).
In this study, the de novo assembly and annotation of the complete genome sequence of C. marina strain JCM 21022T suggested this strain had a circular chromosome that contained a set of crucial genes involved in processes related to surface attachment. In addition, the comparative genomics and phylogenomics analyses revealed genome structural features and allowed us to reconstruct the phylogenetic relationships of species in the family Halomonadaceae.
2 MATERIAL AND METHOD 2.1 DNA extraction, sequencing, and genome assemblyThe C. marina strain JCM 21022T (=DSM 4741T=ATCC 25374T) was obtained from RIKEN Bio Resource Center (BRC) and grown on Marine agar 2216 (BD-Difco). Genomic DNA was extracted using the SDS method combined with RNase A treatment (Wilson, 1997).
Genomic DNA was fragmented, end-repaired, and SMRT bell DNA template libraries (insert size about 10 Kb) were prepared according to the manufacturer's specifications. SMRT sequencing (two SMRT cells) was performed on the Pacific Biosciences RS Ⅱ sequencer using the P4-C2 chemistry according to standard protocols.
Pacbio long reads were obtained from the two SMRT sequencing cells. Reads longer than 1 000 bp with quality values greater than 0.8 were merged. Then the filtered reads were analyzed using the hierarchical genome assembly process (HGAP3) pipeline (Chin et al., 2013). The pipeline uses the longest reads as seeds to recruit all other reads for the construction of highly accurate preassembled reads that were then assembled using the Celera Assembler. To reduce the numbers of remaining insertions or deletions (indels) and base substitution errors in the draft assembly, the Quiver consensus algorithm was used to derive a highly accurate consensus for the final assembly.
2.2 Genome annotationThe location of the replication origins was predicted using the Ori-Finder web service (Gao and Zhang, 2008). Open reading frame (ORF) prediction and annotation were performed using MicroScope, the microbial genome annotation and analysis platform (Vallenet et al., 2013), and verified using Glimmer (Salzberg et al., 1998), RNAmmer (Lagesen et al., 2007) and tRNAscan-SE (Lowe and Eddy, 1997). A graphical circular map of the genome was drawn with Circos v0.66 (Krzywinski et al., 2009).
2.3 Comparative genome analysisWe compared the genome sequences of C. marina JCM 21022T and C. amphilecti KMM 296 (Balabanova et al., 2016a) with BLAST searches (E-value 1.0e-5) to identify similarity at the nucleotide level (Camacho et al., 2009) and with MUMmer 3.23 for a detailed collinearity analysis at the amino acid level (Kurtz et al., 2004). The comparative study on the pan and core genome was performed using BLASTP and SiLiX software (Camacho et al., 2009; Miele et al., 2011).
2.4 Phylogenetic analysesTo elucidate the phylogenetic relationship of C. marina JCM 21022T among species in Halomonadaceae, the concentrated amino acid sequences from 13 species were used to construct a phylogenetic tree. The sequences included 12 complete genomes from Halomonadaceae. The Escherichia coli genome was used to root the tree (Table 1). The sequences were aligned using MAFFT version 5 and adjusted manually (Katoh et al., 2005). All the aligned sites were trimmed using trimAl v1.4 with the options "-automated1" (Capella-Gutiérrez et al., 2009). RAxML61 was employed to reconstruct a maximum likelihood phylogenetic tree for each cluster with an evolutionary model specified as 'PROTGAMMALG' according to PROTTEST-3.4, and a bootstrap significance test was performed with 1 000 replicates (Stamatakis, 2006; Darriba et al., 2011).
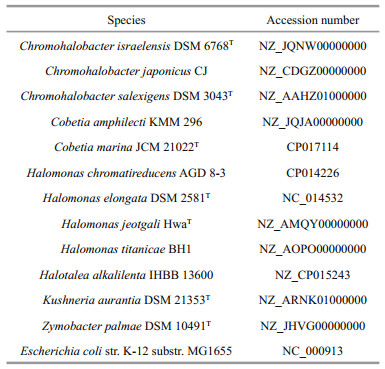
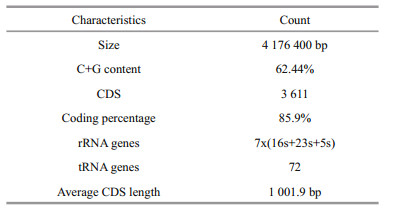
SMRT sequencing of C. marine JCM 21022T generated 224 627 high quality long reads (1 861 380 354 bp) with a coverage of 465×. The HGAP3 pipeline was used to correct sequencing errors. The resulting corrected reads were assembled into a single 4 176 300 bp long contig with an average GC content of 62.44% and 4.07% repeat regions (Fig. 1). The C. marina genome sequence was annotated automatically using the MicroScope platform. A total of 3 611 predicted protein-coding sequences (CDSs) with an average length of 1 002 bp were identified as well as 21 rRNA operons (16S, 23S, and 5S) and 72 predicted tRNAs genes (Table 2). About 33.70% of the open reading frames (ORFs) were predicted to code hypothetical proteins (1 217 ORFs). No plasmids were identified and no clustered regularly interspaced short palindromic repeat (CRISPR) loci were detected in the genome.
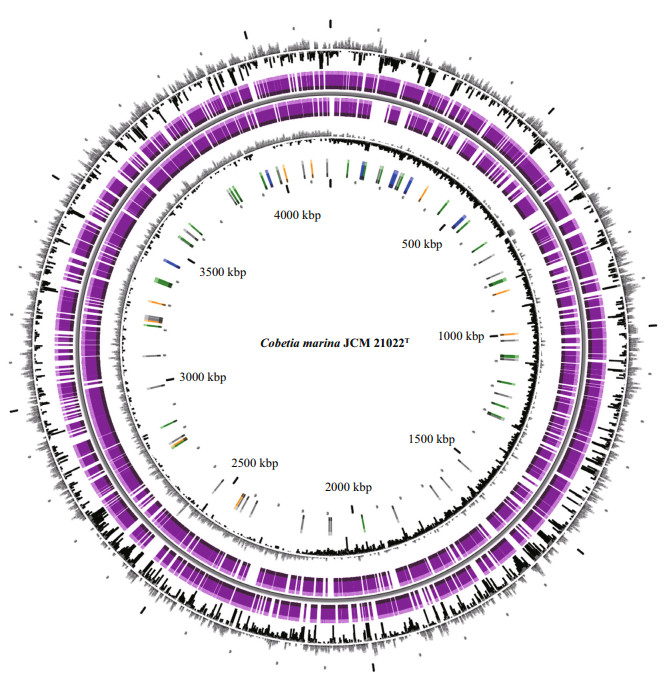
|
| Figure 1 Graphical circular map of the Cobetia marina JCM 21022T chromosome From the outer to inner rings: 1) GC percent deviation (GC window−mean GC content) in a 1 000-bp window; 2) predicted CDSs transcribed in a clockwise direction; 3) predicted CDSs transcribed in a counterclockwise direction; 4) GC skew (G−C/G+C) in a 1 000-bp window; 5) rRNA (blue), tRNA (green), misc_RNA (orange), transposable elements (pink), and pseudogenes (grey). |
The predicted CDSs were annotated with the cluster of orthologous groups (COG) database (Fig. 2). The main annotations were "Amino acid transport and metabolism" (404 genes), "General function prediction only" (471 genes), "Transcription" (260 genes), "Carbohydrate transport and metabolism" (242 genes), "Cell wall/membrane/envelope biogenesis" (220 genes), "Inorganic ion transport and metabolism" (221 genes), and "Translation, ribosomal structure and biogenesis" (183 genes). Further analysis revealed many important genes in the C. marina genome that were related to its ecophysiological status; for example, genes related to cell-surface structures that encoded proteins for curli (Barnhart and Chapman, 2006) and Type IV pili (Spangenberg et al., 1995), and a gene that encoded O-antigen polymerase for extracellular polymer components (Wang and Reeves, 1998). These genes are known to be important for surface attachment in other organisms. In addition, several genes involved in the stress response were also identified in the C. marina genome, including genes coding RpoS, universal stress protein A, stringent starvation proteins (Thomas et al., 2008).
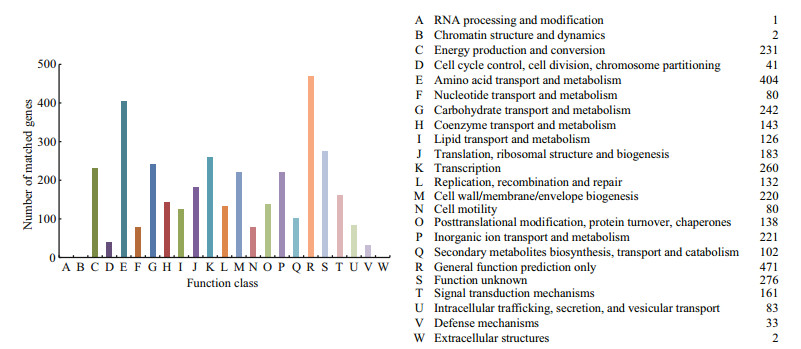
|
| Figure 2 Clusters of orthologous group annotations for the Cobetia marina JCM 21022T genome |
We compared the C. marina JCM 21022T genome with the C. amphilecti KMM 296 genome. The results show only about 67% similarity at the nucleotide level across the two genomes (Fig. 3). The differences were mainly a result of indels in the sequences. The alignment of the two genomes at the amino acid level revealed several chromosomal recombination or transposition and inversions (Fig. 4). A comparative study on the pan and core genes was conducted by comparing the genome sequences of the two strains. As a result, 5 931 core genes (2 646 families) and 7 482 pan genes (4 169 families) were obtained, respectively (Table 3). C. marina JCM 21022T shared 2 962 of 3 611 (82.03%) homologous genes with C. amphilecti KMM 296, implying similar gene functions between them. We detected 645 and 906 strain-specific genes in C. marina JCM 21022T and C. amphilecti KMM 296 respectively.
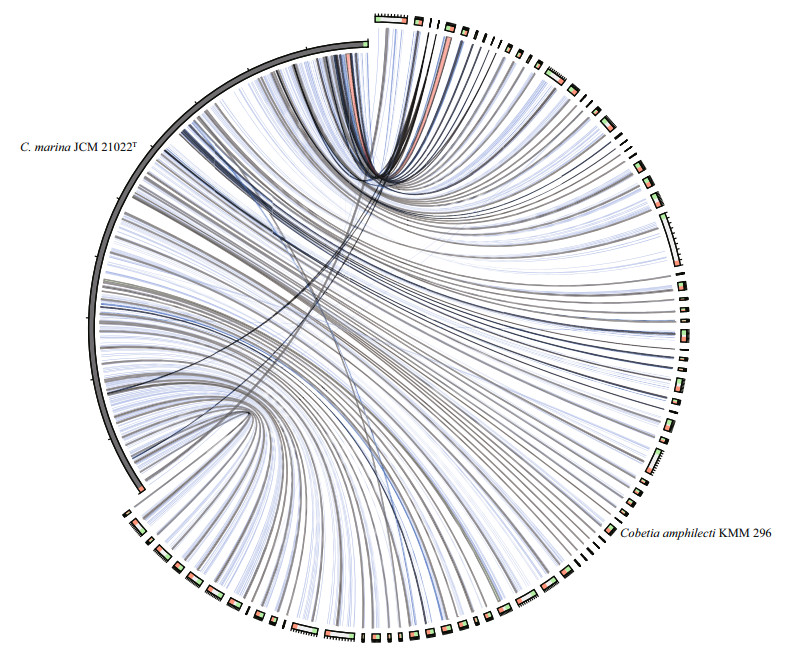
|
| Figure 3 Comparison of the Cobetia marina JCM 21022T and Cobetia amphilecti KMM 296 genomes at the nucleotide level by BLAST search |
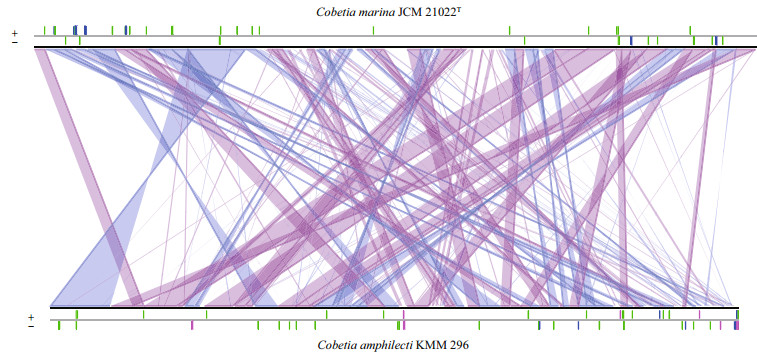
|
| Figure 4 Comparison of the Cobetia marina JCM 21022T and Cobetia amphilecti KMM 296 genomes at the nucleotide level by BLAST search Strand conservations are shown in purple; strand inversions are shown in blue. |

|
To elucidate the evolutionary position of C. marina JCM 21022T in family Halomonadaceae, a data set of 635 concentrated single-copy genes were selected based on the alignment of the genome sequences of 13 related species and its corresponding amino acid sequences were used to construct the phylogenetic tree. The maximum likelihood tree (ML) recovered a well-supported classification among different clades (Fig. 5). The phylogenomic study showed that C. marina JCM 21022T clustered with C. amphilecti KMM 296, indicating a close relationship. The polygenetic tree also showed that species of genus Halomonasclustered together withChromohalobacter, and this cluster then emerged as a sister group to the genus Cobetia. Additionally, the clade uniting Halotalea alkalilenta IHBB 13600 and Zymobacter palmae DSM 10491T was strongly supported by the bootstrap results, and emerged as a sister group to Kushneria aurantia DSM 21353T with high confidence. This group diverged from the other nine species of Halomonadaceae at the base of the phylogenetic tree.
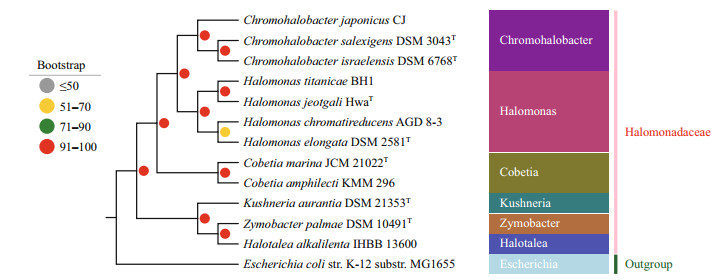
|
| Figure 5 Maximum likelihood phylogenetic tree for Cobetia marina JCM 21022T and the genomes of 12 related species The bootstrap values are for the maximum likelihood with 1 000 replicates. |
In this study, we first presented the whole genomic characteristics of C. marina JCM 21022T, and annotated the predicted genes to several metabolismrelated COG categories. Because C. marina is used as a model organism in marine biofouling studies, its successful attachment on the host surface must be the primary step in biofilm formation (Shea et al., 1995; Arpa Sancet, 2013). As expected, genes that encode for cell-surface structures and extracellular polymer components were detected in the C. marina JCM 21022T genome. The presence of genes that encode curli proteins suggests that C. marina JCM 21022T can attach to invertebrates and algae host surfaces by producing curli, which has been indicated to play significant roles in the interactions between E. coli or Salmonella with plant surfaces (Barak et al., 2005; Jeter and Matthysse, 2005).
In addition, we compared the C. marina JCM 21022T and C. amphilecti KMM 296 (formerly named C. marina KMM 296) genome sequences. Recently, clusters of genes were detected in the C. amphilecti KMM 296 genome that are involved in the transport and metabolism of nitrogen, sulfur, iron, and phosphorus compounds, which have been shown to provide alternative capacity of the inhabitants of the rhizospheres of terrestrial plants, as well as deep-sea ecological communities (Balabanova et al., 2016a). As expected, these genes were also observed in the C. marina JCM 21022T genome. Despite the numerous common genes described above, we identified multiple chromosomal recombination or transposition and inversions between the two genomes. Considering these observations and the relatively large numbers of strain-specific genes, we inferred that there are differences in physiological characteristics and ecological adaptations among C. marina strains, which is consistent with previous studies (Ivanova et al., 2005). Therefore, we consider that it is reasonable to revise the classification of C. marina KMM 296 as C. amphilecti KMM 296 (Balabanova et al., 2016b).
Considering whole genomes can provide more sequence signatures for use in phylogenetic analysis than restricted rDNA sequence, we used 13 bacterial genomes to construct the polygenetic tree of Halomonadaceae. Our results indicated that the genus Halomonas (Vreeland et al., 1980) clustered with Chromohalobacter (Ventosa et al., 1989) first, and then with Cobetia, which was clearly distinguished from the former two groups. Moreover, the phylogenetic relations of other genera such as Kushneria (Sánchez-Porro et al., 2009), Zymobacter (Okamoto et al., 1993), and Halotalea (Ntougias et al., 2007, 2015) were also well resolved. Therefore, we have shown that whole genome information is an appropriate tool in the phylogenetic analysis of bacteria for its ability to provide more evolutionary information than rDNA sequences alone.
In summary, in this study, we sequenced, assembled, and annotated the complete genome sequence of C. marina strain JCM 21022T, and detected a set of crucial genes that may be involved in surface attachment related processes. Although similar gene functions were revealed between C. marina JCM 21022T and C. amphilecti KMM 296 (formerly named C. marina KMM 296), the significant differences identified in the comparative genome analysis resulted from sequence indels and chromosomal recombination. In addition, the phylogenomic study of species in the family Halomonadaceae based on whole genome information was conducted here for the first time, and the relationships among every genus were well resolved. In future studies to comprehensively understand the surface-associated lifestyle of C. marina JCM21022T, genes or gene clusters that endow this species important physiological features should be identified.
| Arahal D R, Castillo A M, Ludwig W, et al, 2002a. Proposal of Cobetia marina gen. nov., comb. nov., within the family Halomonadaceae, to include the species Halomonas marina. Systematic and Applied Microbiology, 25(2): 207–211. Doi: 10.1078/0723-2020-00113 |
| Arahal D R, Ludwig W, Schleifer K H, et al, 2002b. Phylogeny of the family Halomonadaceae based on 23S and 165 rDNA sequence analyses. International Journal of Systematic and Evolutionary Microbiology, 52(1): 241–249. Doi: 10.1099/00207713-52-1-241 |
| Arpa Sancet M P. 2013. Influence of surface properties on adhesion of Cobetia marina and accumulation of marine microfoulers in the ocean. Ruperto Carola University Heidelberg, Heidelberg. |
| Balabanova L A, Golotin V A, Kovalchuk S N, et al, 2016a. The Genome of the marine bacterium Cobetia marina KMM 296 isolated from the mussel Crenomytilus grayanus(Dunker, 1853). Russian Journal of Marine Biology, 42(1): 106–109. Doi: 10.1134/S106307401601003X |
| Balabanova L, Nedashkovskaya O, Podvolotskaya A, et al, 2016b. Data supporting functional diversity of the marine bacterium Cobetia amphilecti KMM 296. Data in Brief, 8: 726–732. Doi: 10.1016/j.dib.2016.06.034 |
| Barak J D, Gorski L, Naraghi-Arani P, et al, 2005. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Applied and Environmental Microbiology, 71(10): 5 685–5 691. Doi: 10.1128/AEM.71.10.5685-5691.2005 |
| Barnhart M M, Chapman M R, 2006. Curli biogenesis and function. Annual Review of Microbiology, 60: 131–147. Doi: 10.1146/annurev.micro.60.080805.142106 |
| Baumann L, Baumann P, Mandel M, et al, 1972. Taxonomy of aerobic marine eubacteria. Journal of Bacteriology, 110(1): 402–429. |
| Baumann L, Bowditch R D, Baumann P, 1983. Description of Deleya gen. nov. created to accommodate the marine species Alcaligenes aestus, A. pacificus, A. cupidus, A. venustus, and Pseudomonas marina. International Journal of Systematic and Evolutionary Microbiology, 33(4): 793–802. |
| Camacho C, Coulouris G, Avagyan V, et al, 2009. BLAST+:architecture and applications. BMC Bioinformatics, 10: 421. Doi: 10.1186/1471-2105-10-421 |
| Capella-Gutiérrez S, Silla-Martínez J M, Gabaldón T, 2009. trimAl:a tool for automated alignment trimming in largescale phylogenetic analyses. Bioinformatics, 25(15): 1 972–1 973. Doi: 10.1093/bioinformatics/btp348 |
| Chin C S, Alexander D H, Marks P, et al, 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature Methods, 10(6): 563–569. Doi: 10.1038/nmeth.2474 |
| Cobet A B, Wirsen C Jr, Jones G E, 1970. The effect of nickel on a marine bacterium, Arthrobacter marinus sp. nov. Journal of General Microbiology, 62(2): 159–169. Doi: 10.1099/00221287-62-2-159 |
| Copeland A, O'Connor K, Lucas S, et al, 2011. Complete genome sequence of the halophilic and highly halotolerant Chromohalobacter salexigens type strain (1H11T). Standards in Genomic Sciences, 5(3): 379–388. Doi: 10.4056/sigs.2285059 |
| Darriba D, Taboada G L, Doallo R, et al, 2011. ProtTest 3:fast selection of best-fit models of protein evolution. Bioinformatics, 27(8): 1 164–1 165. Doi: 10.1093/bioinformatics/btr088 |
| de la Haba R R, Arahal D R, Márquez M C, et al, 2010. Phylogenetic relationships within the family Halomonadaceae based on comparative 23S and 16S rRNA gene sequence analysis. International Journal of Systematic and Evolutionary Microbiology, 60(4): 737–748. Doi: 10.1099/ijs.0.013979-0 |
| Dobson S J, Franzmann P D. 1996. Unification of the genera Deleya (Baumann et al. 1983), Halomonas (Vreeland et al. 1980), and Halovibrio (Fendrich 1988) and the species Paracoccus halodenitrificans (Robinson and Gibbons 1952) into a single genus, Halomonas, and placement of the genus Zymobacter in the family Halomonadaceae. International Journal of Systematic Bacteriology, 46(2): 550-558. |
| Eid J, Fehr A, Gray J, et al, 2009. Real-time DNA sequencing from single polymerase molecules. Science, 323(5910): 133–138. Doi: 10.1126/science.1162986 |
| Gao F, Zhang C T, 2008. Ori-Finder:a web-based system for finding oriC s in unannotated bacterial genomes. BMC Bioinformatics, 9: 79. Doi: 10.1186/1471-2105-9-79 |
| Ista L K, Fan H Y, Baca O, et al, 1996. Attachment of bacteria to model solid surfaces:oligo(ethylene glycol) surfaces inhibit bacterial attachment. FEMS Microbiology Letters, 142(1): 59–63. Doi: 10.1111/fml.1996.142.issue-1 |
| Ista L K, Pérez-Luna V H, López G P, 1999. Surface-grafted, environmentally sensitive polymers for biofilm release. Applied and Environmental Microbiology, 65(4): 1 603–1 609. |
| Ivanova E P, Christen R, Sawabe T, et al, 2005. Presence of ecophysiologically diverse populations within Cobetia marina strains isolated from marine invertebrate, algae and the environments. Microbes and Environments, 20(4): 200–207. Doi: 10.1264/jsme2.20.200 |
| Jeter C, Matthysse A G, 2005. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Molecular Plant-Microbe Interactions, 18(11): 1 235–1 242. Doi: 10.1094/MPMI-18-1235 |
| Katoh K, Kuma K I, Toh H, et al, 2005. MAFFT version 5:improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33(2): 511–518. Doi: 10.1093/nar/gki198 |
| Krzywinski M, Schein J, Birol I, et al, 2009. Circos:an information aesthetic for comparative genomics. Genome Research, 19(9): 1 639–1 645. Doi: 10.1101/gr.092759.109 |
| Kurtz S, Phillippy A, Delcher A L, et al, 2004. Versatile and open software for comparing large genomes. Genome Biology, 5(2): R12. Doi: 10.1186/gb-2004-5-2-r12 |
| Lagesen K, Hallin P, Rødland E A, et al, 2007. RNAmmer:consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research, 35(9): 3 100–3 108. Doi: 10.1093/nar/gkm160 |
| Lowe T M, Eddy S R, 1997. tRNAscan-SE:a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 25(5): 955–964. Doi: 10.1093/nar/25.5.0955 |
| Mata J A, Martínez-Cánovas J, Quesada E, et al, 2002. A detailed phenotypic characterisation of the type strains of Halomonas species. Systematic and Applied Microbiology, 25(3): 360–375. Doi: 10.1078/0723-2020-00122 |
| Miele V, Penel S, Duret L, 2011. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics, 12: 116. Doi: 10.1186/1471-2105-12-116 |
| Ntougias S, Lapidus A, Copeland A, et al, 2015. High-quality permanent draft genome sequence of the extremely osmotolerant diphenol degrading bacterium Halotalea alkalilenta AW-7T, and emended description of the genus Halotalea. Standards in Genomic Sciences, 10: 52. Doi: 10.1186/s40793-015-0052-7 |
| Ntougias S, Zervakis G I, Fasseas C. 2007. Halotalea alkalilenta gen. nov., sp. nov., a novel osmotolerant and alkalitolerant bacterium from alkaline olive mill wastes, and emended description of the family Halomonadaceae Franzmann et al. 1989, emend. Dobson and Franzmann 1996. International Journal of Systematic and Evolutionary Microbiology, 57(9): 1 975-1 983. |
| Okamoto T, Taguchi H, Nakamura K, et al, 1993. Zymobacter palmae gen. nov., sp. nov., a new ethanol-fermenting peritrichous bacterium isolated from palm sap. Archives of Microbiology, 160(5): 333–337. |
| Roberts R J, Carneiro M O, Schatz M C, 2013. The advantages of SMRT sequencing. Genome biology, 14: 405. Doi: 10.1186/gb-2013-14-6-405 |
| Romanenko L A, Tanaka N, Svetashev V I, et al, 2013. Description of Cobetia amphilecti sp. nov., classification of Halomonas halodurans as a later heterotypic synonym of Cobetia marina and emended descriptions of the genus Cobetia and Cobetia marina. International Journal of Systematic and Evolutionary Microbiology, 63(1): 288–297. |
| Salzberg S L, Delcher A L, Kasif S, et al, 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Research, 26(2): 544–548. Doi: 10.1093/nar/26.2.544 |
| Sánchez-Porro C, de la Haba R R, Cruz-Hernández N, et al, 2013. Draft Genome of the marine Gammaproteobacterium Halomonas titanicae. Genome Announcements, 1(2): e00083–13. |
| Sánchez-Porro C, de la Haba R R, Soto-Ramírez N, et al, 2009. Description of Kushneria aurantia gen. nov., sp. nov., a novel member of the family Halomonadaceae, and a proposal for reclassification of Halomonas marisflavi as Kushneria marisflavi comb. nov., of Halomonas indalinina as Kushneria indalinina comb. nov. and of Halomonas avicenniae as Kushneria avicenniae comb. nov. International Journal of Systematic and Evolutionary Microbiology, 59(2): 397–405. Doi: 10.1099/ijs.0.001461-0 |
| Schwibbert K, Marin-Sanguino A, Bagyan I, et al, 2011. A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581T. Environmental Microbiology, 13(8): 1 973–1 994. Doi: 10.1111/j.1462-2920.2010.02336.x |
| Sharko F S, Shapovalova A A, Tsygankova S V, et al, 2016. Draft genome sequence of "Halomonaschromatireducens" Strain AGD 8-3, a Haloalkaliphilic Chromate-and Selenite-Reducing Gammaproteobacterium. Genome Announcements, 4(2): e00160–16. |
| Shea C, Lovelace L J, Smith-Somerville H E, 1995. Deleya marina as a model organism for studies of bacterial colonization and biofilm formation. Journal of Industrial Microbiology, 15(4): 290–296. Doi: 10.1007/BF01569982 |
| Spangenberg C, Fislage R, Sierralta W, et al, 1995. Comparison of type Ⅳ-pilin genes of Pseudomonas aeruginosa of various habitats has uncovered a novel unusual sequence. FEMS Microbiology Letters, 125(2-3): 265–273. Doi: 10.1111/fml.1995.125.issue-2-3 |
| Stamatakis A, 2006. RAxML-Ⅵ-HPC:maximum likelihoodbased phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22(21): 2 688–2 690. Doi: 10.1093/bioinformatics/btl446 |
| Thomas T, Evans F F, Schleheck D, et al, 2008. Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS One, 3(9): e3252. Doi: 10.1371/journal.pone.0003252 |
| Vallenet D, Belda E, Calteau A, et al, 2013. MicroScope-an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Research, 41(D1): D636–D647. Doi: 10.1093/nar/gks1194 |
| Ventosa A, Gutierrez M C, Garcia M T, et al, 1989. Classification of "Chromobacterium marismortui" in a new genus, Chromohalobacter gen. nov., as Chromohalobacter marismortui comb. nov., nom. rev. International Journal of Systematic Bacteriology, 39(4): 382–386. Doi: 10.1099/00207713-39-4-382 |
| Vreeland R H, Litchfield C D, Martin E L, et al, 1980. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. International Journal of Systematic Bacteriology, 30(2): 485–495. Doi: 10.1099/00207713-30-2-485 |
| Wang L, Reeves P R, 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infection and Immunity, 66(8): 3 545–3 551. |
| Wilson K. 1997. Preparation of genomic DNA from bacteria. In: Ausubel F M, Bent R, Kingston R E et al eds. Current Protocols in Molecular Biology. John Wiley & Sons, Inc., New York. p. 2. 4. 1-2. 4. 5. |
 2018, Vol. 36
2018, Vol. 36