Institute of Oceanology, Chinese Academy of Sciences
Article Information
- XIA Mengjie(夏梦洁), PEI Feng(裴峰), MU Changkao(母昌考), YE Yangfang(叶央芳), WANG Chunlin(王春琳)
- Disruption of bacterial balance in the gut of Portunus trituberculatus induced by Vibrio alginolyticus infection
- Chinese Journal of Oceanology and Limnology, 36(5): 1891-1898
- http://dx.doi.org/10.1007/s00343-018-7121-x
Article History
- Received Jun. 2, 2017
- accepted in principle Aug. 8, 2017
- accepted for publication Sep. 13, 2018
2 Collaborative Innovation Center for Zhejiang Marine High-Efficiency and Healthy Aquaculture, Ningbo 315211, China
The swimming crab, Portunus trituberculatus, is one of the most important fishery crab species and widely cultured on a commercial scale in China. However, the diseases due to bacterial infection frequently occur in farmed swimming crabs. Numerous pathogens have been reported so far in P. trituberculatus including Pseudomonas putida (Wang et al., 2007), Vibrio alginolyticus (Liu et al., 2007), V. parahaemolyticus (Yan et al., 2010), V. metschnikovii (Wan et al., 2011), V. harveyi (Zhang et al., 2014), and V. natriegens (Bi et al., 2016). Among them, V. alginolyticus is regarded as the main causative pathogen (Liu et al., 2007). Many studies have focused on the effects of V. alginolyticus infection on swimming crabs at the levels of gene, transcript, protein, and metabolite. For instance, the sequences of a cascade of immune-related genes such as PtHsp70 gene (Cui et al., 2010), PtToll gene (Li et al., 2015), and PtSRB gene (Yang et al., 2016) in swimming crabs have been cloned and their transcriptional expressions are closely associated with V. alginolyticus challenge. Furthermore, at the level of metabolite, an acute V. alginolyticus infection induces the tissue-specific metabolomic alterations in swimming crabs (Ye et al., 2016).
Gut is an essential organ which has important physiological functions in P. trituberculatus. Trillions of microbes, collectively known as the microbiota, inhabit the gut and are intricately linked to host's health (Kau et al., 2011). In humans, a body of evidence has elucidated that gut microbiota plays a fundamental role in nutrient absorption and normal immune function for the benefit of host health (Brestoff and Artis, 2013; Song et al., 2015). Disruption to its normal balance may contribute to pathological conditions, such as inflammatory bowel disease (Dicksved et al., 2008), obesity (Tremaroli and Bäckhed, 2012), and liver cirrhosis (Qin et al., 2014). For crustaceans, gut microbiota is widely involved in the organ development (Cheung et al., 2015), nutrition (Xiong et al., 2015), immunity (Wu et al., 2014), and diseases (Olmos et al., 2011). In turn, the habitat, health, and diets of crustaceans such as crabs are responsible for shaping the symbiotic gut bacteria pattern (Wu et al., 2015; Zhang et al., 2016). Although a close relationship between crabs and their gut microbiota is increasingly being accepted, little is known about the influences of vibrio infection on gut bacteria of P. trituberculatus. Unraveling the alteration in bacterial community composition in response to vibrio infection is crucial to better understanding the pathogenesis and progression of V. alginolyticus-infected disease.
An amplicon sequencing method ought to be a suitable choice for defining the dynamic bacterial change in swimming crab gut responding to V. alginolyticus infection. This is because amplicon sequencing enables systemic detection of bacterial composition in biological system and dynamic responses to both endogenous and exogenous perturbation (Qin et al., 2010). Numerous examples have illustrated that the amplicon sequencing method is powerful to reveal the gut bacterial community structure of crustaceans such as crabs (Rungrassamee et al., 2016; Zhang et al., 2016) and shrimps (Xiong et al., 2015, 2017; Zhu et al., 2016).
In the current study, 16S rRNA gene sequencing was used to investigate temporal alterations of the gut bacterial community composition in P. trituberculatus in response to 72-h V. alginolyticus challenge. Our aim is to understand the vibrio-induced dynamic changes in gut bacterial community during the development of vibriosis of P. trituberculatus.
2 MATERIAL AND METHOD 2.1 Crab breeding and V. alginolyticus infectionMale swimming crabs with a weight of approximately 100 g of each were collected from a local aquaculture farm (Ninghai, China) and cultured in an aerated recycling seawater with a salinity of 24±1 at room temperature for one week of acclimatization. Crabs were fed with clam meat once daily at 17:00–18:00. Cultural seawater was changed in the next morning.
For V. alginolyticus infection experiment, three vigorous crabs were randomly collected and used as 0 h control group. Nine crabs were infected with 10 μL/g live V. alginolyticus resuspended in 0.01 mol/L phosphate buffered saline (PBS, pH 7.4, 107 cfu/mL) via arthrodial membrane of the last walking leg of each crab. These crabs were used as the infected ones and cultured in a cement pond (8 m×3 m×2 m, length×width×depth). Other nine crabs which received an injection of 10 μL/g PBS were served as uninfected controls and cultured in another cement pond. Three infected and three uninfected crabs were separately collected from these two ponds at time points of 24, 48, and 72 h, respectively. Each collected crab was kept into ice seawater for hypothermic anesthesia and then sacrificed for gut tissue including its contents. Each crab gut sample was snap-frozen with liquid nitrogen and stored at -80℃ until further analysis.
2.2 DNA extraction, 16S rRNA gene amplification, and Illumina MiSeq sequencingGut samples from 0, 24, 48, and 72 h of postinfection were employed for bacterial community composition analysis using Illumina MiSeq sequencing technology as described previously (Caporaso et al., 2012). The genomic DNA was extracted from approximately 300 mg gut tissue including its contents using a PowerFecal™ DNA Isolation kit (MO BIO, USA). The obtained DNA extracts were quantified using a Qubit 2.0 fluorometer (Life Technologies, USA) and submitted to Shanghai Majorbio Bio-Pharm Technology Co., Ltd. for 16S rRNA gene amplification, library preparation, and 250 bp paired-end Illumina MiSeq sequencing. In brief, the dual-indexed bacterial/archaeal primer pair 515F (5'-GTGCCAGCMGCCGCGGTAA-3') and 806R (5'-GGACTACNVGGGTWTCTAAT-3') containing adaptor sequences for the MiSeq platform (Caporaso et al., 2012) was used to amplify the 16S rRNA gene V4 region.
An aliquot of 10 ng of purified DNA template from each sample was amplified in triplicate in a 30 μL reaction system (denaturation 98℃ for 1 min, followed by 35 cycles of denaturation at 98℃ for 10 s, annealing at 50℃ for 30 s, and extension at 72℃ for 30 s, with a final extension at 72℃ for 5 min). The resultant PCR products were pooled together for minimizing the reaction-level PCR bias. The purified PCR products were subsequently quantified using the Qubit 2.0 fluorometer. An equimolar amount of PCR amplicons was combined into one pooled sample and sequenced on the Illumina MiSeq platform.
2.3 16S amplicon processing and analysisThe sequences generated in this study were deposited in the Sequence Read Archive of DDBJ (http://www.ddbj.nig.ac.jp) and are available under accession number DRA005812. Raw FASTQ files were demultiplexed with QIIME (v1.8.0) (Caporaso et al., 2010a). The paired reads were assembled with the FLASH using the default settings (Magoč and Salzberg, 2011) and subsequently filtered using QIIME. In brief, the reads were truncated at any site containing more than three sequential bases receiving a Phred quality score less than 20. The reads containing ambiguous base calls were discarded. Also, the reads with less than 75% (of the total read length) consecutive high quality base calls were discarded. The remaining sequences were chimera assessed using USEARCH (Edgar et al., 2011). Following filtering the chimera reads, the sequences were clustered into operational taxonomic units (OTUs) at 97% nucleotide similarity level using the pick_open_ reference_otus.py script. The most abundant sequences in the OTUs were assigned against the RDP database (Release 11.3) and aligned using PyNAST (Caporaso et al., 2010b). The OTUs assigned to Archaea, chloroplasts, and unclassified reads were discarded prior to subsequent analysis. The full dataset (n=21) contained 758 725 clean reads (mean 36 130 reads per sample). The alpha-diversity (Shannon index) and richness of observed species were calculated by even rarefication at 10 000 reads per sample using QIIME. The principal coordinates analysis (PCoA) plot based on Bray-Curtis distance was used to visualize the sample clusters and dissimilarity in bacterial community composition between groups. The obtained averaged relative abundances of bacteria were subjected to classical statistical analysis of Student's t-test, and the P values less than 0.05 were considered statistically significant with *.
3 RESULT 3.1 Alpha-diversity of bacteriaThe α-diversity of gut bacterial composition reflected as obtained OTU numbers and Shannon indices was lower in vibrio-infected groups during the earlier 48 h whereas higher at 72 h than in controls (Fig. 1). However, no significant difference in bacterial α-diversity was observed between the infection and control groups.
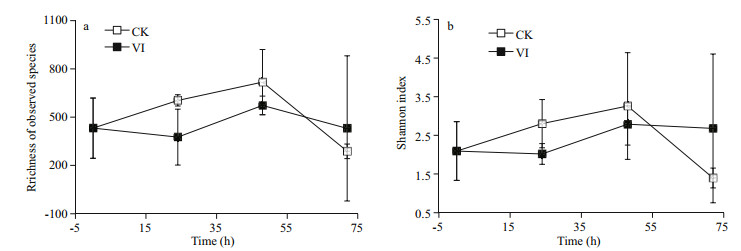
|
| Figure 1 The changes of richness (a) and diversity (b) of gut bacteria of swimming crabs after Vibrio alginolyticus infection CK: non-infected control crabs; VI: vibrio-infected crabs. |
To obtain an overview of the effects of V. alginolyticus infection on crab gut bacterial community structure, PCoA of the bacterial community composition from gut samples of swimming crabs was constructed based on the first two principle components (PC1 and PC2) (Fig. 2). The PCoA plot (Fig. 2a) showed that V. alginolyticus infection induced a dissimilarity in the bacterial composition of swimming crab gut. The samples of infection groups displayed more discrete than control groups and showed an infection time-dependent pattern. Thus, the bacterial community structure of crab gut was changed by V. alginolyticus infection. Furthermore, the bacterial communities with relative abundance > 1% (Fig. 2b) made a greater contribution than those with relative abundance < 1% (Fig. 2c).
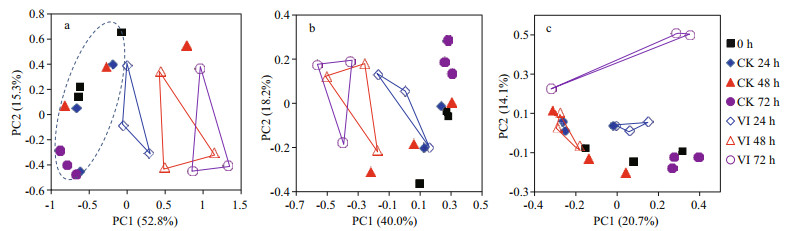
|
| Figure 2 Principal coordinates analysis (PCoA) based on Bray-Curtis distance for gut bacterial community composition from swimming crabs after Vibrio alginolyticus infection a. all of bacteria; b. the bacteria with relative abundance > 1%; c. the bacteria with relative abundance < 1%. 0 h: non-infected control crabs at 0 h; CK 24 h: non-infected control crabs at 24 h; CK 48 h: non-infected control crabs at 48 h; CK 72 h: non-infected control crabs at 72 h; VI 24 h: vibrioinfected crabs at 24 h; VI 48 h: vibrio-infected crabs at 48 h; VI 72 h: vibrio-infected crabs at 72 h. |
To further reveal how a bacterial community composition was changed by vibrio infection, the relative abundance for the main bacterial phyla in crab gut was analyzed. Our results showed that more than 97% of the clean sequences were classified at the phylum level (Fig. 3). Proteobacteria, Fusobacteria, Tenericutes, and Bacteroidetes were dominant microbial divisions with the fluctuated abundance in both the vibrio-infected crabs and controls at all time points. At the phylum level, vibrio-infected crabs at 24 h had lower abundance of Bacteroidetes, Cyanobacteria, Saccharibacteria, and Hydrogenedentes than its control counterpart (Fig. 3). Vibrio-infected crabs at 48 h had lower abundance of Saccharibacteria and Hyd24-12 than its control counterpart (Fig. 3b). Vibrio-infected crabs at 72 h had significant higher abundance of Proteobacteria but lower abundance of Fusobacteria than its control counterpart (Fig. 3a).

|
| Figure 3 The averaged relative abundance of bacteria at the phylum level in the gut of swimming crabs after Vibrio alginolyticus infection a. the bacteria with relative abundance > 1%; b. the significantly changed bacteria with relative abundance < 1%. *: P < 0.05, by Student's t-test, the infected crab group vs its control counterpart; red and green asterisks denoted the significantly increased and decreased abundance of bacterial population in the infected crab group compared with its control counterpart, respectively. Asterisks were marked on the control group in plot b for easy marking. The meaning of the abbreviation of group name is given in Fig. 2. |
At the genus level, the bacterial community was also different between the vibrio-infected crabs and their control counterparts at the different time points of post-infection (Figs. 4, 5). Compared to 24 h control counterpart, vibrio-infected crabs at 24 h had lower abundance of Pseudoalteromonas, Alteromonas, Carboxylicivirga, Draconibacterium, Marinicella, Muricauda, Neptunomonas, Lewinella, Portibacter, Hellea, Arenicella, SM1A02, Pseudoteredinibacter, Kordiimonas, Candidatus_Thiobios, Candidatus_Hepatoplasma, unclassified genera in Saprospiraceae, Hyphomonadaceae, Phyllobacteriaceae, Chloroflexi, Alcaligenaceae, and Burkholderiales, some norank bacteria in Cyanobacteria, GR-WP33-58, as well as some uncultured genera in Cytophagaceae (Fig. 5). Vibrioinfected crabs at 48 h had higher abundance of Vibrio, Wenyingzhuangia, Thalassotalea, Lishizhenia, Hahella, Sneathiella, unclassified genera in Cellvibrionales, and some uncultured genera in Chitinophagaceae and Xanthomonadaceae but lower abundance of Chryseobacterium, and some norank bacteria in Hyd24-12 when compared to its control counterpart (Figs. 4, 5). Furthermore, vibrio-infected crabs at 72 h only had lower abundance of Photobacterium and some uncultured genera in Leptotrichiaceae when compared to its control counterpart (Figs. 4, 5). In particular, Vibrio was continuously increased over the whole infection time in the infected crabs whereas not in control crabs (Fig. 4b).
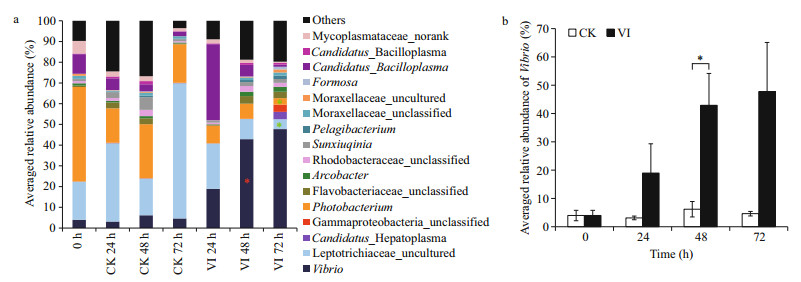
|
| Figure 4 The averaged relative abundance of bacteria with relative abundance > 1% at the genus level (a) and Vibrio (b) in the gut of swimming crabs after Vibrio alginolyticus infection The meaning of the abbreviation of group name is given in Fig. 2. The meaning of the asterisks is given in Fig. 3. |
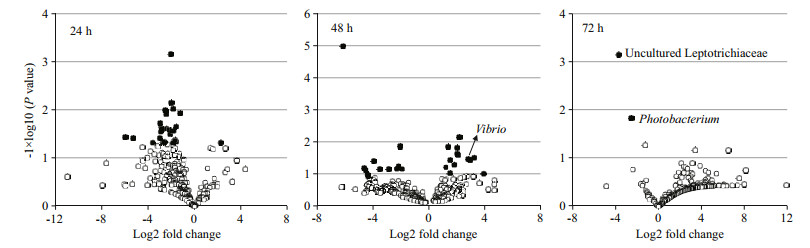
|
| Figure 5 Volcano plot of abundance fold change data of vibrio-induced gut bacteria in swimming crabs For the volcano plots, bacterial abundance log2 fold change between the vibrio-infected crabs and the non-infected control crabs is plotted on the x axis, and the P value for a t-test of differences between the vibrio-infected crabs and the non-infected control crabs (-1×log10 scale) is plotted on the y axis. Each circle represents one bacterial species at the genus level. Empty circles, P > 0.05; filled circles, P < 0.05. |
Our study shows that only the proportion of the phylum Proteobacteria in the crab gut displays a continuous increase after the vibrio infection. Within this phylum, the abundances of more than 15 genera display a significant change whereas Vibrio overwhelmingly dominates the crab gut from 48 h of post-infection. Thus, the increased proportion of Proteobacteria should attribute to the increased abundance of Vibrio (OTU349). Vibrio spp. are commonly found and commensal in the gut of marine animals such as crabs (Chen et al., 2015), shrimps (Liu et al., 2004) and fishes (Ohwada et al., 1980). Overabundance of certain bacterial population may trigger the transition of gut bacteria from a healthy, sub-optimal to a diseased status (Xiong et al., 2015). For instance, the increased abundance of Rhodobacteraceae spp., Vibrio spp. and Flavobacteriaceae spp. in the gut of shrimp has been observed to be parallel with the disease progression (Xiong et al., 2017). In this study, Photobacterium is predominant in the non-infected swimming crabs, which is the same as the previous observation in the gut of healthy swimming crabs (Zeng et al., 2016). However, Vibrio overwhelms the normal dominant bacteria in the crab gut after vibrio infection. Vibrio comprises a catalog of species, many of which are commensals. However, a catalog of species are pathogens which have already been found in P. trituberculatus including V. alginolyticus (Liu et al., 2007), V. parahaemolyticus (Yan et al., 2010), V. metschnikovii (Wan et al., 2011), V. harveyi (Zhang et al., 2014), and V. natriegens (Bi et al., 2016). In this study, OTU349 likely shares 99% similarity with V. alginolyticus based on the Basic Local Alignment Search Tool results. If so, this notorious pathogen not only successfully colonizes but also becomes overabundant in the gut of swimming crab, strongly indicating a close relationship with the vibriosis of swimming crabs.
4.2 The disrupted bacterial balance in crab gutThe homeostasis of intestinal bacteria in animal host is very important to maintain the host health and resilience against pathogens (De Schryver and Vadstein, 2014). Although no significant difference in bacterial α-diversity was induced by V. alginolyticus infection, a more discrete variation of bacterial community structure of vibrio-infected crab gut at each time point than that of controls indicates that the normal balance of the gut bacteria is disrupted by the pathogen invasion. The population of the gut bacteria is modulated following vibrio infection. Such a modulation is manifested not only in the members with the high relative abundance but also ones with the low relative abundance. Obviously, the deviation of gut bacterial community structure mainly attributes to the bacterial members with the relative abundance > 1%, which probably playing more important role than those with low relative abundance. However, it is also worthy to note that the significantly changed bacterial members with the relative abundance < 1% were more than ones with the relative abundance > 1% and an obvious deviation of gut bacterial community structure was resulted from the bacterial members with the low abundance at the later period of infection (72 h). In fact, the bacteria with low relative abundance also exhibit important biological functions and could be noted when many of them are significantly changed. For instance, the species of Pseudoalteromonas (Yoshikawa et al., 1997) and Alteromonas (Gauthier, 1976) have anti-bacterial activities, which might be useful to resist the survival and colonization of exogenous V. alginolyticus. However, a significant reduction in the abundances of Pseudoalteromonas and Alteromonas obviously ameliorates the competition stress of V. alginolyticus for nutrients and living space. Therefore, such a dysbiosis of crab gut bacteria may add pathological infection and accelerate disease progression. This notion is consistent with a recent study in which the balance of intestinal bacterial population plays a crucial role in the mortality rates of two shrimp species after V. harveyi exposure (Rungrassamee et al., 2016). The black tiger shrimp with the dysbiosis of intestinal bacterial population has a higher mortality rate than the Pacific white shrimp which can restore the bacterial balance. Such a view on the balance of gut bacteria is also concordant with the occurrence of human diseases. It is unbalanced microbial population that results in various diseases such as obesity (Turnbaugh et al., 2009), cardiovascular disease (Wang et al., 2011), liver cirrhosis (Qin et al., 2014), and Parkinson's disease (Sampson et al., 2016). Therefore, tracking the deviation of gut bacterial community composition from healthy one could provide a novel insight into disease warning and diagnosis. However, how these changed bacteria especially Vibrio contribute to vibriosis progression is not yet known and requires to be validated in the further work.
5 CONCLUSIONTo sum up, the microbiomic analyses revealed that V. alginolyticus infection resulted in dynamic changes of the gut bacteria in swimming crabs. Such changes were highlighted by the overwhelming overabundance of Vibrio and a significant fluctuation in the gut bacteria including the bacteria with high relative abundance and especially those with low relative abundance. These findings reveal that crab vibriosis gradually develops with the infection time of V. alginolyticus and tightly relates to the dysbiosis of gut bacterial community composition. The importance of the balance of gut bacterial community structure in the progression of swimming crab disease needs more attention.
Bi K R, Zhang X J, Yan B L, Gao H, Gao X J, Sun J J. 2016. Isolation and molecular identification of Vibrio natriegens from diseased Portunus trituberculatus in China. Journal of the World Aquaculture Society, 47(6): 854-861.
DOI:10.1111/jwas.2016.47.issue-6 |
Brestoff J R, Artis D. 2013. Commensal bacteria at the interface of host metabolism and the immune system. Nature Immunology, 14(7): 676-684.
DOI:10.1038/ni.2640 |
Caporaso J G, Bittinger K, Bushman F D, DeSantis T Z, Andersen G L, Knight R. 2010b. PyNAST:a flexible tool for aligning sequences to a template alignment. Bioinformatics, 26(2): 266-267.
DOI:10.1093/bioinformatics/btp636 |
Caporaso J G, Kuczynski J, Stombaugh J, Bittinger K, Bushman F D, Costello E K, Fierer N, Peña A G, Goodrich J K, Gordon J I, Huttley G A, Kelley S T, Knights D, Koenig J E, Ley R E, Lozupone C A, McDonald D, Muegge B D, Pirrung M, Reeder J, Sevinsky J R, Turnbaugh P J, Walters W A, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010a. QⅡME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5): 335-336.
DOI:10.1038/nmeth.f.303 |
Caporaso J G, Lauber C L, Walters W A, Berg-Lyons D, Huntley J, Fierer N, Owens S M, Betley J, Fraser L, Bauer M, Gormley N, Gilbert J A, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal, 6(8): 1621-1624.
DOI:10.1038/ismej.2012.8 |
Chen X B, Di P P, Wang H M, Li B L, Pan Y J, Yan S L, Wang Y J. 2015. Bacterial community associated with the intestinal tract of Chinese mitten crab (Eriocheir sinensis)farmed in Lake Tai, China. PLoS One, 10(4): e0123990.
DOI:10.1371/journal.pone.0123990 |
Cheung M K, Yip H Y, Nong W, Law P T W, Chu K H, Kwan H S, Hui J H L. 2015. Rapid change of microbiota diversity in the gut but not the hepatopancreas during gonadal development of the new shrimp model Neocaridina denticulata. Marine Biotechnology, 17(6): 811-819.
DOI:10.1007/s10126-015-9662-8 |
Cui Z X, Liu Y, Luan W S, Li Q Q, Wu D H, Wang S Y. 2010. Molecular cloning and characterization of a heat shock protein 70 gene in swimming crab (Portunus trituberculatus). Fish & Shellfish Immunology, 28(1): 56-64.
|
De Schryver P, Vadstein O. 2014. Ecological theory as a foundation to control pathogenic invasion in aquaculture. The ISME Journal, 8: 2360-2368.
DOI:10.1038/ismej.2014.84 |
Dicksved J, Halfvarson J, Rosenquist M, Järnerot G, Tysk C, Apajalahti J, Engstrand L, Jansson J K. 2008. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. The ISME Journal, 2: 716-727.
DOI:10.1038/ismej.2008.37 |
Edgar R C, Haas B J, Clemente J C, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16): 2194-2200.
DOI:10.1093/bioinformatics/btr381 |
Gauthier M J. 1976. Alteromonas rubra sp. nov., a new marine antibiotic-producing bacterium. International Journal of Systematic Bacteriology, 26: 459-466.
DOI:10.1099/00207713-26-4-459 |
Kau A L, Ahern P P, Griffin N W, Goodman A L, Gordon J I. 2011. Human nutrition, the gut microbiome and the immune system. Nature, 474(7351): 327-336.
DOI:10.1038/nature10213 |
Li M, Li C W, Wang J F, Song S Q. 2015. Molecular characterization and expression of a novel Toll gene from the swimming crab Portunus trituberculatus. Molecular Immunology, 67(2 Pt B): 388-397.
|
Liu C H, Cheng W, Hsu J P, Chen J C. 2004. Vibrio alginolyticus infection in the white shrimp Litopenaeus vannamei confirmed by polymerase chain reaction and 16S rDNA sequencing. Diseases of Aquatic Organisms, 61(1-2): 169-174.
|
Liu Q, Li H Y, Wang Q, Liu P, Dai F Y, Li J. 2007. Identification and phylogenetic analysis of a strain of Vibrio alginolyticus, a pathogen in Portunus trituberculatus with toothpaste disease. Marine Freshwater Research, 28(4): 9-13.
(in Chinese with English abstract) |
Magoč T, Salzberg S L. 2011. FLASH:fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27(21): 2957-2963.
DOI:10.1093/bioinformatics/btr507 |
Ohwada K, Tabor P S, Colwell R R. 1980. Species composition and barotolerance of gut microflora of deep-sea benthic macrofauna collected at various depths in the Atlantic ocean. Applied and Environmental Microbiology, 40(4): 746-755.
|
Olmos J, Ochoa L, Paniagua-Michel J, Contreras R. 2011. Functional feed assessment on Litopenaeus vannamei using 100% fish meal replacement by soybean meal, high levels of complex carbohydrates and Bacillus probiotic strains. Marine Drugs, 9(6): 1119-1132.
DOI:10.3390/md9061119 |
Qin J J, Li R Q, Raes J, Arumugam M, Burgdorf K S, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende D R, Li J H, Xu J M, Li S C, Li D F, Cao J J, Wang B, Liang H Q, Zheng H S, Xie Y L, Tap J, Lepage P, Bertalan M, Batto J M, Hansen T, Le Paslier D, Linneberg A, Nielsen H B, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H M, Yu C, Li S T, Jian M, Zhou Y, Li Y R, Zhang X Q, Li S G, Qin N, Yang H M, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich S D, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464(7285): 59-65.
DOI:10.1038/nature08821 |
Qin N, Yang F L, Li A, Prifti E, Chen Y F, Shao L, Guo J, Le Chatelier E, Yao J, Wu L J, Zhou J W, Ni S J, Liu L, Pons N, Batto J M, Kennedy S P, Leonard P, Yuan C H, Ding W C, Chen Y T, Hu X J, Zheng B W, Qian G R, Xu W, Ehrlich S D, Zheng S S, Li L J. 2014. Alterations of the human gut microbiome in liver cirrhosis. Nature, 513(7516): 59-64.
DOI:10.1038/nature13568 |
Rungrassamee W, Klanchui A, Maibunkaew S, Karoonuthaisiri N. 2016. Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. Journal of Invertebrate Pathology, 133: 12-19.
DOI:10.1016/j.jip.2015.11.004 |
Sampson T R, Debelius J W, Thron T, Janssen S, Shastri G G, Ilhan Z E, Challis C, Schretter C E, Rocha S, Gradinaru V, Chesselet M F, Keshavarzian A, Shannon K M, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian S K. 2016. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell, 167(6): 1469-1480.e12.
DOI:10.1016/j.cell.2016.11.018 |
Song X Y, Dai D, He X, Zhu S, Yao Y K, Gao H C, Wang J J, Qu F F, Qiu J, Wang H L, Li X X, Shen N, Qian Y C. 2015. Growth factor FGF2 cooperates with interleukin-17 to repair intestinal epithelial damage. Immunity, 43(3): 488-501.
DOI:10.1016/j.immuni.2015.06.024 |
Tremaroli V, Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nature, 489(7415): 242-249.
DOI:10.1038/nature11552 |
Turnbaugh P J, Hamady M, Yatsunenko T, Cantarel B L, Duncan A, Ley R E, Sogin M L, Jones W J, Roe B A, Affourtit J P, Egholm M, Henrissat B, Heath A C, Knight R, Gordon J I. 2009. A core gut microbiome in obese and lean twins. Nature, 457(7228): 480-484.
DOI:10.1038/nature07540 |
Wan X H, Shen H, Wang L B, Cheng Y X. 2011. Isolation and characterization of Vibrio metschnikovii causing infection in farmed Portunus trituberculatus in China. Aquaculture International, 19(2): 351-359.
DOI:10.1007/s10499-011-9422-3 |
Wang G X, Huang Z R, Yuan M. 2007. Isolation and identification of the pathogen of milk disease cultured in swimming crab Portunus tritubercularus (portunidae). Journal of Northwest A & F University (Natural Science Edition), 35(6): 29-33.
(in Chinese with English abstract) |
Wang Z N, Klipfell E, Bennett B J, Koeth R, Levison B S, DuGar B, Feldstein A E, Britt E B, Fu X M, Chung Y M, Wu Y P, Schauer P, Smith J D, Allayee H, Tang W H W, DiDonato J A, Lusis A J, Hazen S L. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature, 472(7341): 57-63.
DOI:10.1038/nature09922 |
Wu H J, Sun L B, Li C B, Li Z Z, Zhang Z, Wen X B, Hu Z, Zhang Y L, Li S K. 2014. Enhancement of the immune response and protection against Vibrio parahaemolyticus by indigenous probiotic Bacillus strains in mud crab (Scylla paramamosain). Fish & Shellfish Immunology, 41: 156-162.
|
Wu T, Zhang Z L, Cai C F, Ye Y T, Zhu J M, Li T. 2015. The effect of pectin and xylan on intestinal microflora structure of Chinese mitten crab. Genomics and Applied Biology, 34(4): 745-753.
(in Chinese with English abstract) |
Xiong J B, Wang K, Wu J F, Qiuqian L, Yang K J, Qian Y X, Zhang D M. 2015. Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Applied Microbiology and Biotechnology, 99(16): 6911-6919.
DOI:10.1007/s00253-015-6632-z |
Xiong J B, Zhu J Y, Dai W F, Dong C M, Qiu Q F, Li C H. 2017. Integrating gut microbiota immaturity and diseasediscriminatory taxa to diagnose the initiation and severity of shrimp disease. Environmental Microbiology, 19(4): 1490-1501.
DOI:10.1111/1462-2920.13701 |
Yan B L, Qin G M, Bao Z H, Zhang X J, Bi K R, Qin L. 2010. Isolation and identification of Vibrio parahaemolyticus from diseased Portunus trituberculatus L. Marine Science Bulletin, 29(5): 560-566.
(in Chinese with English abstract) |
Yang N, Zhang D F, Tao Z, Li M, Zhou S M, Wang G L. 2016. Identification of a novel class B scavenger receptor homologue in Portunus trituberculatus:molecular cloning and microbial ligand binding. Fish & Shellfish Immunology, 58: 73-81.
|
Ye Y F, Xia M J, Mu C K, Li R H, Wang C L. 2016. Acute metabolic response of Portunus trituberculatus to Vibrio alginolyticus infection. Aquaculture, 463: 201-208.
DOI:10.1016/j.aquaculture.2016.05.041 |
Yoshikawa K, Takadera T, Adachi K, Nishijima M, Sano H. 1997. Korormicin, a novel antibiotic specifically active against marine Gram-negative bacteria, produced by a marine bacterium. The Journal of Antibiotics, 50(11): 949-953.
DOI:10.7164/antibiotics.50.949 |
Zeng T L, Ye Y F, Mu C K, Wang K, Li R H, Wang C L. 2016. Gut microbiota and metabolic phenotype of Portunus trituberculatus. Chinese Journal of Analytical Chemistry, 44(12): 1867-1873.
DOI:10.1016/S1872-2040(16)60978-7 |
Zhang M L, Sun Y H, Chen L Q, Cai C F, Qiao F, Du Z Y, Li E C. 2016. Symbiotic bacteria in gills and guts of Chinese mitten crab (Eriocheir sinensis) differ from the free-living bacteria in water. PLoS One, 11(1): e0148135.
DOI:10.1371/journal.pone.0148135 |
Zhang X J, Bai X S, Yan B L, Bi K R, Qin L. 2014. Vibrio harveyi as a causative agent of mass mortalities of megalopa in the seed production of swimming crab Portunus trituberculatus. Aquaculture International, 22(2): 661-672.
DOI:10.1007/s10499-013-9695-9 |
Zhu J Y, Dai W F, Qiu Q F, Dong C M, Zhang J J, Xiong J B. 2016. Contrasting ecological processes and functional compositions between intestinal bacterial community in healthy and diseased shrimp. Microbial Ecology, 72(4): 975-985.
DOI:10.1007/s00248-016-0831-8 |
 2018, Vol. 36
2018, Vol. 36


