Institute of Oceanology, Chinese Academy of Sciences
Article Information
- SONG Wei(宋炜), ZHU Yefei(朱叶飞), WANG Lumin(王鲁民), JIANG Keji(蒋科技), ZHANG Fengying(张凤英), MA Chunyan(马春艳), MA Lingbo(马凌波)
- Identification and profiling of microRNAs of Euphausia superba using Illumina deep sequencing
- Chinese Journal of Oceanology and Limnology, 36(6): 2278-2287
- http://dx.doi.org/10.1007/s00343-019-7229-7
Article History
- Received Aug. 4, 2017
- accepted in principle Oct. 31, 2017
- accepted for publication Jan. 2, 2018
2 Key Laboratory of East China Sea Fishery Resources Exploitation, Ministry of Agriculture, East China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Shanghai 200090, China
MicroRNAs (miRNAs) are an abundant class of conserved, approximately 18–25 nucleotides in length, endogenous non-coding RNA that regulate target gene expression through cleavage or translational inhibition (Cai et al., 2004), and then affecting crucial biological and metabolic processes. At least 35 828 miRNAs from hundreds of species, are recorded in the online repository miRBase 21 (Kozomara and Griffiths-Jones, 2014). Though there is near-perfect pairing in plant miRNAs, animal miRNAs regulate target genes through imperfect sequence-specific binding to the 3' or 5' untranslated regions (Jopling et al., 2005) or even to the coding regions (Tay et al., 2008; Ryan et al., 2010), causing translational repression and, in certain cases, degradation of the mRNA (He and Hannon, 2004). Furthermore, regulation form of miRNA is not always inhibited, as the expression of tumor necrosis factor-alpha was up-regulated by miRNA-369-3p in human cells (Vasudevan et al., 2007). It is generally considered that miRNAs play the role of regulator at the post-transcriptional level. However, recent research has found that miR-24-1 activated enhancer RNA expression, altered histone modification, and increased the enrichment of p300 and RNA polymerase Ⅱ at the enhancer locus (Xiao et al., 2017). Increasing evidence indicates that miRNAs are involved in embryogenesis and organogenesis (Melton et al., 2010; Pauli et al., 2011), cell development and migration, immune response (Brown et al., 2007; O'Connell et al., 2009), and hormone secretion (Cortez et al, 2011). Moreover, alternative expression of miRNAs can lead to series diseases or defective development (Calin et al, 2002).
High-throughput sequencing allows rapid and sensitive detection of miRNAs and can facilitate the discovery of novel, low abundance, and species-or tissue-specific miRNAs (Hafner et al., 2008). Using both deep sequencing and bioinformatics analysis to identify miRNAs in white shrimp Litopenaeus vannamei and giant freshwater prawn Macrobrachium rosenbergii, hundreds of conserved miRNAs were obtained (Tan et al., 2013; Xi et al., 2015). Antarctic krill Euphausia superba is possibly the world's most abundant animal species, providing a massive protein resource that is targeted in the fisheries of numerous countries. However, with climate change and fishing operations, have resulted in a steady decline of the population density of Antarctic krill in the past decades (Atkinson et al., 2004; Cascella et al., 2015). For the sustainable development of this species, genomic and transcriptomic researches are expected to improve our understanding of the physiology and biology of krill.
This study aimed to identify and profile miRNA in the eyestalk and muscle tissues of Antarctic krill. The eyestalk is a complex and major organ for the secretion of hormones and involved in osmotic regulation, molting, epidermal color patterns, osmoregulation, modulation of glycaemia and reproduction (Turner et al., 2013; Katayama, 2016). The results will be useful to future research on the function of miRNAs involved in regulating development in E. superba.
2 MATERIAL AND METHOD 2.1 Sample collectionHealthy adult krill Euphausia superba from Antarctic waters of the Southern Ocean were collected on-board the Chinese icebreaking research vessel Snow Dragon and then transported to the laboratory. The eyestalk and muscle tissues were promptly dissected, added to RNAfixer, and store at -80℃ until RNA extraction.
2.2 Construction of small RNA library and Illumina deep sequencingTotal RNA was extracted using TRIzol® reagent (Invitrogen, CA, USA) following the manufacturer's protocol. Total RNA quantity and purity were calculated with a 2100 Bioanalyzer and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number >7.0. Approximately 2 μg of purified total RNA was used to construct a small RNA library, according to the protocol of the NEBNext® Small RNA Library Prep Set for Illumina® (New England Biolabs, USA). Thereafter, the concentration and quality of small RNA librariy were measured using a Qubit® 2.0 Fluorometer (Life Technologies) and an Agilent 2100 Bioanalyzer (Agilent Technologies). Finally, we performed the single-end sequencing (35 bp) on the Illumina HiSeq 2500 platform.
2.3 Bioinformatics analysisRemoving adaptors sequences, low quality tags, and sequences containing polyA tails allowed us to get clean reads and summarize the size distribution of small RNA. The retained 17–35 nt reads were mapped onto the reference transcriptome, filtering out unmapped sequences. The mapped reads were then classified into the tRNA, Repeat and Rfam database to remove rRNA, tRNA, snRNA snoRNA and other ncRNA, as well as the repeat sequence. Next, the sequences were mapped to intron and exon to identify degradation fragment of mRNA. After series modification, the retained sequences were blast to the miRNA precursor and mature miRNA of all animals in miRBase without mismatch, in order to identify conserved miRNAs.
MicroRNA read counts were normalized to the individual lane size by dividing each read count by the total number of reads in million per lane. Differentially expressed miRNAs between the two libraries were discovered using the R package DEGseq. P-values were calculated using DEGseq software. Scatter plots were generated based on log2 normalized read counts, with fold change = log2 (normalized read counts in sample1/normalized read counts in sample2). Statistical significance was set at a P-value of ≤0.05 and fold change ≥2. To predict the targets of the miRNAs, we used sRNA-tools of target module.
2.4 Quantitative Real Time-PCRTo validate and characterize the differentially expressed miRNAs identified in muscle and eyestalk of krill, we performed quantitative real-time PCR (qRT-PCR) busing the miRNA specific stem-loop primer approach (Table 1). Real-time PCR was performed using a standard SRYB Green PCR Kit protocol on an Applied Biosystems 7500 Sequence Detection System (P/N: 4329002, Applied Biosystems, Foster City, CA). The 20 μL PCR volume included 1 μL of RT product, 10 μL of 2X SYBR Green qPCR Master Mix, and 1 μL of primer (5 mmol/L each of the forward and reverse primers). The reactions were incubated at 95℃ for 5 min, followed by 40 cycles of 95℃ for 15 s, 60℃ for 40 s. The relative expression of each miRNA was determined by comparative Ct (ΔΔCt) methods using U6 as the internal reference gene. Each experiment was repeated independently at least three times.
Statistical analysis was performed using SPSS v19.0 software. Data are presented as mean±standard error (S.E.). Statistical significance was determined using one-way analysis of variance (ANOVA) and post-hoc Duncan multiple range tests. Significance was set at P < 0.05.
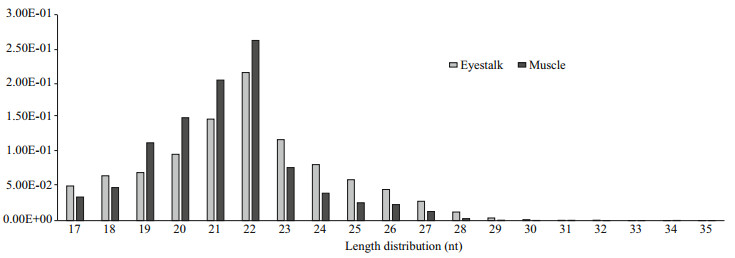
3 RESULT 3.1 General Features of the Solexa sequencing of small RNAsThis study generated two distinct small RNA libraries from the eyestalk (E) and muscle (M) to identify the Antarctic krill miRNAs using deep sequencing (Illumina Inc.). We obtained 21 609 615 and 24 755 663 raw reads from the E and M small RNA libraries, respectively. After filtering the junk reads, adapter sequences and the sequences outside the range of 18 to 35 nt, 19 304 586 reads from the E group and 23 005 104 reads from the M group were obtained for analysis (Table 2). Afterwards, we analyzed the length distribution of these clean reads to evaluate the sequencing quality. The length distribution of these clean sequence reads showed a peak at 20–24 nt for both libraries (Fig. 1), consistent with the mainly products of Dicer derived cleavage (Yao et al., 2010).
|
| Fig.1 Length distributions of the high quality filtered sequencing reads in the small RNA library of eyestalk and muscle for Antarctic krill |
Since the whole genomic sequence of the krill is unavailable, we analyzed the small RNAs from both libraries were analyzed using the de novo transcriptome from gill and hepatopancreas tissues of E. superba as the reference (unpublished data). Using Bowtie 2 v2.1.0 (Langmead and Salzberg 2012), 63.93% of the clean reads for the E group and 34.37% for the M group could be mapped to the de novo transcriptome of E. superba without any mismatches. Clean reads that could not be mapped to krill transcriptome, and matched sequence that did not form a hairpin structure, were removed. The mapped sequences were further categorized and annotated by aligning with sequences in Rfam and Repeat database (Fig. 2). More than 80% of the mapping reads were identified as the unannotated (unano) group, while the smaller portion of the other RNAs lacked a genome resource. The repeat associated sequences from the E library were more various than those from the M library (Table 3).

|
| Fig.2 Annotation and classification of the small RNAs in the (a) eyestalk and (b) muscle library |
To investigate conserved miRNAs in eyestalk (E) and muscle (M) of E. superba, we aligned the mapped sequences with a non-redundant reference set of all animal miRNAs registered in miRBase 21 by allowing at most two mismatches outside of the seed region. The matched Small RNAs were identified as conserved miRNA orthologs in E. superba. We obtained 139 478 reads for E and 215 525 reads for M from the mapped reads as known miRNAs. Most of the predicted E. superba mature miRNA sequences, from 20 to 24 nt, for the E library started with an adenine residue at their 5′ end, which is consistent with findings for the aphid Acyrthosiphon pisum (Legeai et al., 2010) and the trematode Schistosoma mansoni (De Souza Gomes et al., 2011), whereas the M library started with a thymidine residue. Ultimately, 236 conserved mature miRNAs, which contained 64 miRNA-5p and 66 miRNA-3p, were identified from the two libraries, and 48 of them were co-expressed in both libraries.
After normalizing the individual mapped reads, the most abundant miRNAs identified in each library were listed in in descending order of abundance (Tables 4, 5); these results indicated that the expression of several of the abundant miRNAs may be tissue-specific. The most abundant of the known miRNA was miR-750, with 92 583 reads detected in muscle, with an average value of 68. Among the small RNA library of eyestalk, miR-1304-3p was the most accumulated of the known miRNAs, and showed 56 386 reads, with an average value of 105. Especially, miR-277a was abundantly expressed, with more than 10 000 reads in both libraries.
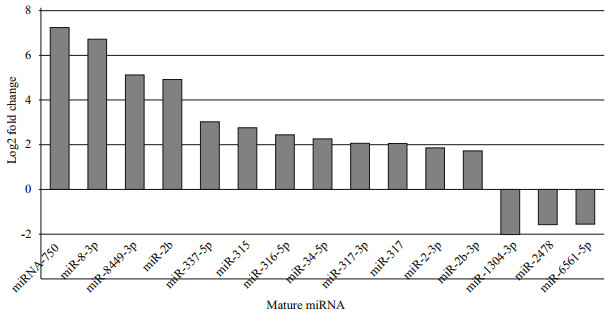
We compared the miRNA expression levels between the two small RNA libraries of eyestalk and muscle tissues. For this purpose, 56 mature miRNAs were significantly up/down regulated in terms of expression fold change, based on a thresholds of 2-times fold change (FC) and an overall P-value of < 0.05. Among the most abundant miRNAs that were expressed in relatively high amounts in the two libraries (Fig. 3), miR-1304-3p (-2 log2 FC), miR-2478 and miR-6561-5p (both -1.5 log2 FC), were significantly down regulated, whereas miR-750, miR-1b and miR-8-3p (all ~7 log2 FC), were significantly up regulated compared to eyestalk. Interestingly, the expression of three miR-1 members (miR-1a, -1b, and -1-3p) was significantly higher in muscle than in eyestalk, showing muscle-biased expression.
|
| Fig.3 Differential expression of known miRNAs between eyestalk and muscle Normalized reads ≥100. |
To confirm the accuracy of the sequencing results, four miRNAs were selected and determined by qRTPCR via miRNA specific stem-loop primers (Fig. 4). Among the four miRNAs, miR-1a and miR-1304-3p were relatively high expressed in muscle and eyestalk respectively, and miR-317-3p and miR-184 were coexpressed in the two organs. The relative expression levels of these miRNAs were consistent with the results of Solexa sequencing.

|
| Fig.4 Comparison of the expression levels of four miRNAs in eyestalk (E), muscle (M) and hepatopancreas (H) discovered through qRT-PCR The asterisks indicate that the differences between the mean value are statistically significant (P < 0.05) compared to hepatopancreas. |
The increasing evidence demonstrates that miRNAs as an abundant group of fundamental regulators, participating in many biological and phsiological processes (Begemann, 2008). In the last decades, over thirty thousand of miRNA have been identified in various species (miRBase v21). Improvements in RNA deep sequencing technologies and bioinformatics have improved detection of miRNA transcriptome detection in crustacean, which are lacking a reference genome, such as in freshwater crayfish Procambarus clarkia (Ou et al., 2013), mud crab Scylla paramamosain (Li et al., 2013), mitten crab Eriocheir sinensis (Song et al., 2014), and tadpole shrimp Triops cancriformis (Ikeda et al., 2015). However, no data are available regarding miRNAs of krill. In this study, were constructed two distinct small RNA libraries of from the eyestalk (E) and muscle (M) tissues, to identify the Antarctic krill miRNAs by high-throughput sequencing technologies. We obtained 19 304 586 and 23 005 104 clean reads from the small RNA libraries of E and M, respectively. The length distribution analysis revealed a peak at the range of 20–24 nt for both libraries (Fig. 1), which matched to the typical length products for Dicer derived cleavage (Yao et al., 2010). Using the mRNA, Rfam and Repeat databases, more than 80% of the mapped reads were classified as the unannotated (unano) group, and the other small RNAs accounted for the smaller proportion. These results suggested the limitations of the reference transcriptome and the high quality of these new libraries.
In this study, a total of 236 conserved miRNAs from eyestalk and muscle tissues were identified and annotated. We then focused on the most abundantly expressed and conserved miRNAs in the the E. superba libraries (Tables 3, 4). Additional sequencing is required for identification of low expression level and novel miRNAs. The most abundant known miRNAs in the libraries were miR-750, with 92 583 reads in muscle, and miR-1304-3p, with 56 386 reads in eyestalk, while the average values were less than 106, revealing a wide range of different expression levels. The polymorphic distribution of miRNAs often suggests a role in distinct tissues or biological processes. As shown in previous research, miR-317, miR-2, miR-1175 and miR-277 are evolutionary conserved miRNAs, having similar sequences and functions among species (Ruby et al., 2007). The miR-317 is involved in sex peptides reverse (Fricke et al., 2014) and and possibly targets Drosophila cyclin B during the G2-M transition (Pushpavalli et al., 2014). The miR-2 demonstrated high expression in the first metaphase (MI) of meiosis and luciferase reporter gene assay analysis showed that miR-2 can down regulate the crab cyclin B gene (Song et al., 2014). In addition, miR-2 has a potential function in neurodevelopment in the Drosophila (Marco et al., 2012). The miR-1175 is regarded as key regulator of quinone oxidoreductase and glutathione peroxidase, which are important to the immune system (Ou et al., 2013). The expression level of miR-277-3p in the goldenrod gall fly Eurosta solidaginis was significantly reduced in frozen larvae (Courteau et al., 2012). The Vimar enzyme is employed as an indicator of mitochondrial oxidative capacity and was revealed to be under the regulation of miR-277 (Bruce, et al., 2004; Bos, 2005). In this study, maybe due to the environment change during the krill catches, miR-277a was found to be a dominant miRNA, detected in both muscle and eyestalk, which suggests that miR-277 might be involved in freeze tolerance and energy metabolism in the Antarctic krill.
We further compared the transcriptional levels of the miRNAs between the two small RNA libraries of eyestalk and muscle tissues. In terms of differently expressed miRNAs between the libraries, 34 miRNAs were up regulated and 22 miRNAs were down regulated in muscle as compared with eyestalk. Notably, miR-750 was the most abundant miRNA of this study, by more than 7-fold in muscle as compared to eyestalk. In addition, all the three members of the miR-1 family showed muscle-biased expression. As demonstrated by previous study (Huang et al., 2014), miRNAs mediate cell proliferation and differentiation of muscles by modulating muscle development related gene expression. Conditional knockout (cKO) of Dicer1 (a distinct protein that participate in the biosynthesis of miRNAs) from skeletal muscle leaded to myofibers hypoplasia and apoptosis in mouse (O'Rourke et al., 2007). It was found that miR-1 is restrictive or abundantly expressed in skeletal muscle tissue, which suggests an important role in regulating muscle development and growth (Yan et al., 2012). Moreover, in zebrafish, actin organization and sarcomere assembly were disrupted under defective miR-1 expression (Mishima et al., 2009). Analogously, loss-function of miR-1 in Drosophila lead to serious amyotrophy (Kwon et al., 2005; Sokol and Ambros, 2005). MHCs (myosin heavy chains), a major component of myosins, were identified as a potential growth marker gene (Harrington and Rodgers, 1984). In the swimming crab Portunus trituberculatus, miR-750 has a higher expression level in larger-sized individuals than in smaller individuals, and MHC was the only predicted target gene of miR-750, indicating that it may stimulate growth by regulating MHC expression (Ren et al., 2016). Overall, the miR-750 and miR-1family are the key candidates involved in krill muscle development.
We found that the two most abundant miRNAs (miR-1304-3p and miR-6561-5p) were significantly up regulated in eyestalk as compared with those in muscle. Intriguingly, both of those miRNAs were also new candidates detected in embryonic stem cells (Morin et al., 2008; Shao et al., 2012), signifying their involvement in cell differentiation, migration and apoptosis., although the available information on miRNAs in crustaceans is quickly expanding, most research have focused on the immune response (Li et al., 2013), responses to viral infection (Li et al., 2013) and stress responses (Lv et al., 2016), as detected mainly in hepatopancreas, gills and gonad tissues. However, few studies have considered the expression patterns and functions of miRNAs in eyestalk of crustacean. This is the first profiling of miRNAs in krill, and the research has identified hundreds of miRNAs in eyestalk and muscle, some of these miRNAs displaing a potential role in the immune response, reproduction, energy metabolism and muscle development. These data provide a theoretical basis for the functional research of miRNA in E. superba.
5 CONCLUSIONTo our knowledge, this is the first study on miRNA expression profiles of eyestalk and muscle tissues in krill. A total of 236 distinct miRNAs were identified in the small RNA libraries from eyestalk and muscle of the krill. Differential expression analysis demonstrates that some of the identified miRNAs are tissue-specific or tissue-biased, which suggested potential roles in the regulation of endocrine function, growth and development of krill. Specially, miR-750 and miR-1 were found to be highly expressed in krill muscle, revealing a significant role in krill muscle development. The results presented here will be of assistance in miRNAs functional studies of E. superba, such as aspects of the species' in immune response, reproduction, energy metabolism, and muscle development.
6 DATA AVAILABILITY STATEMENTThe datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
7 ACKNOWLEDGMENTWe thank Liwen Bianji, Edanz Group China (www. liwenbianji.cn), for editing the English text of a draft of this manuscript.
Atkinson A, Siegel V, Pakhomov E, Rothery P. 2004. Longterm decline in krill stock and increase in salps within the southern Ocean. Nature, 432(7013): 100-103.
DOI:10.1038/nature02996 |
Begemann G. 2008. MicroRNAs and RNA interference in zebrafish development. Zebrafish, 5(2): 111-119.
DOI:10.1089/zeb.2008.0528 |
Bos J L. 2005. Linking Rap to cell adhesion. Current Opinion in Cell Biology, 17(2): 123-128.
DOI:10.1016/j.ceb.2005.02.009 |
Brown B D, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. 2007. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nature Biotechnology, 25(12): 1457-1467.
DOI:10.1038/nbt1372 |
Bruce C R, Kriketos A D, Cooney G J, Hawley J A. 2004. Disassociation of muscle triglyceride content and insulin sensitivity after exercise training in patients with Type 2 diabetes. Diabetologia, 47(1): 23-30.
DOI:10.1007/s00125-003-1265-7 |
Cai X Z, Hagedorn C H, Cullen B R. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA, 10(12): 1957-1966.
DOI:10.1261/rna.7135204 |
Calin G A, Dumitru C D, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce C M. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America, 99(24): 15524-15529.
DOI:10.1073/pnas.242606799 |
Cascella K, Jollivet D, Papot C, Léger N, Corre E, Ravaux J, Clark M S, Toullec J Y. 2015. Diversification, evolution and sub-functionalization of 70kDa heat-shock proteins in two sister species of antarctic krill:differences in thermal habitats, responses and implications under climate change. PLoS One, 10(4): e0121642.
DOI:10.1371/journal.pone.0121642 |
Cortez M A, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood A K, Calin G A. 2011. MicroRNAs in body fluidsthe mix of hormones and biomarkers. Nature Reviews Clinical Oncology, 8(8): 467-477.
DOI:10.1038/nrclinonc.2011.76 |
Courteau L A, Storey K B, Morin P Jr. 2012. Differential expression of microRNA species in a freeze tolerant insect, Eurosta solidaginis. Cryobiology, 65(3): 210-214.
DOI:10.1016/j.cryobiol.2012.06.005 |
De Souza Gomes M, Muniyappa M K, Carvalho S G, GuerraSá R, Spillane C. 2011. Genome-wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni. Genomics, 98(2): 96-111.
DOI:10.1016/j.ygeno.2011.05.007 |
Fricke C, Green D, Smith D, Dalmay T, Chapman T. 2014. MicroRNAs influence reproductive responses by females to male sex peptide in Drosophila melanogaster. Genetics, 198(4): 1603-1619.
DOI:10.1534/genetics.114.167320 |
Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. 2008. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods, 44(1): 3-12.
DOI:10.1016/j.ymeth.2007.09.009 |
Harrington W F, Rodgers M E. 1984. Myosin. Annual Review of Biochemistry, 53: 35-73.
DOI:10.1146/annurev.bi.53.070184.000343 |
He L, Hannon G J. 2004. MicroRNAs:small RNAs with a big role in gene regulation. Nature Reviews Genetics, 5(7): 522-531.
DOI:10.1038/nrg1379 |
Huang T Z, Cui Y L, Zhang X B. 2014. Involvement of viral microRNA in the regulation of antiviral apoptosis in shrimp. Journal of Virology, 88(5): 2544-2554.
DOI:10.1128/JVI.03575-13 |
Ikeda K T, Hirose Y, Hiraoka K, Noro E, Fujishima K, Tomita M, Kanai A. 2015. Identification, expression, and molecular evolution of microRNAs in the "living fossil" Triops cancriformis (tadpole shrimp). RNA, 21(2): 230-242.
DOI:10.1261/rna.045799.114 |
Jopling C L, Yi M, Lancaster A M, Lemon S M, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science, 309(5740): 1577-1581.
DOI:10.1126/science.1113329 |
Katayama H. 2016. Structure-activity relationship of crustacean peptide hormones. Bioscience, Biotechnology, and Biochemistry, 80(4): 633-641.
DOI:10.1080/09168451.2015.1116932 |
Kozomara A, Griffiths-Jones S. 2014. miRBase:annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 42(Database issue): D68-D73.
DOI:10.1093/nar/gkt1181 |
Kwon C, Han Z, Olson E N, Srivastava D. 2005. MicroRNA1influences cardiac differentiation in Drosophila and regulates Notch signaling. Proceedings of the National Academy of Sciences of the United States of America, 102(52): 18986-18991.
DOI:10.1073/pnas.0509535102 |
Langmead B, Salzberg S L. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods, 9(4): 357-359.
DOI:10.1038/nmeth.1923 |
Legeai F, Rizk G, Walsh T, Edwards O, Gordon K, Lavenier D, Leterme N, Méreau A, Nicolas J, Tagu D, JaubertPossamai S. 2010. Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genomics, 11: 281.
DOI:10.1186/1471-2164-11-281 |
Li S K, Zhu S, Li C B, Zhang Z, Zhou L Z, Wang S J, Wang S Q, Zhang Y L, Wen X B. 2013. Characterization of microRNAs in mud crab Scylla paramamosain under Vibrio parahaemolyticus infection. PLoS One, 8(8): e73392.
DOI:10.1371/journal.pone.0073392 |
Lv J J, Liu P, Gao B Q, Li J. 2016. The identification and characteristics of salinity-related microRNAs in gills of Portunus trituberculatus. Cell Stress and Chaperones, 21(1): 63-74.
DOI:10.1007/s12192-015-0641-9 |
Marco A, Hooks K, Griffiths-Jones S. 2012. Evolution and function of the extended miR-2 microRNA family. RNA Biology, 9(3): 242-248.
DOI:10.4161/rna.19160 |
Melton C, Judson R L, Blelloch R. 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature, 463(7281): 621-626.
DOI:10.1038/nature08725 |
Mishima Y, Abreu-Goodger C, Staton A A, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright A J, Giraldez A J. 2009. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes & Development, 23(5): 619-632.
DOI:10.1101/gad.1760209 |
Morin R D, O'Connor M D, Griffith M, Kuchenbauer F, Delaney A, Prabhu A L, Zhao Y J, McDonald H, Zeng T, Hirst M, Eaves C J, Marra M A. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Research, 18(4): 610-621.
DOI:10.1101/gr.7179508 |
O'Connell R M, Chaudhuri A A, Rao D S, Baltimore D. 2009. Inositol phosphatase SHIP1 is a primary target of miR-155. Proceedings of the National Academy of Sciences of the United States of America, 106(17): 7113-7118.
DOI:10.1073/pnas.0902636106 |
O'Rourke J R, Georges S A, Seay H R, Tapscott S J, McManus M T, Goldhamer D J, Swanson M S, Harfe B D. 2007. Essential role for Dicer during skeletal muscle development. Developmental Biology, 311(2): 359-368.
DOI:10.1016/j.ydbio.2007.08.032 |
Ou J T, Li Y, Ding Z F, Xiu Y J, Wu T, Du J, Li W J, Zhu H X, Ren Q, Gu W, Wang W. 2013. Transcriptome-wide identification and characterization of the Procambarus clarkii microRNAs potentially related to immunity against Spiroplasma eriocheiris infection. Fish & Shellfish Immunology, 35(2): 607-617.
DOI:10.1016/j.fsi.2013.05.013 |
Pauli A, Rinn J L, Schier A F. 2011. Non-coding RNAs as regulators of embryogenesis. Nature Reviews Genetics, 12(2): 136-149.
DOI:10.1038/nrg2904 |
Pushpavalli S N C V L, Sarkar A, Bag I, Hunt C R, Ramaiah M J, Pandita T K, Bhadra U, Pal-Bhadra M. 2014. Argonaute-1 functions as a mitotic regulator by controlling Cyclin B during Drosophila early embryogenesis. The FASEB Journal, 28(2): 655-666.
DOI:10.1096/fj.13-231167 |
Ren X Y, Cui Y T, Gao B Q, Liu P, Li J. 2016. Identification and profiling of growth-related microRNAs of the swimming crab Portunus trituberculatus by using Solexa deep sequencing. Marine Genomics, 28: 113-120.
DOI:10.1016/j.margen.2016.03.010 |
Ruby J G, Stark A, Johnston W K, Kellis M, Bartel D P, Lai E C. 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Research, 17(12): 1850-1864.
DOI:10.1101/gr.6597907 |
Ryan B M, Robles A I, Harris C C. 2010. Genetic variation in microRNA networks:the implications for cancer research. Nature Reviews Cancer, 10(6): 389-402.
DOI:10.1038/nrc2867 |
Shao P, Liao J Y, Guan D G, Yang J H, Zheng L L, Jing Q, Zhou H, Qu L H. 2012. Drastic expression change of transposon-derived piRNA-like RNAs and microRNAs in early stages of chicken embryos implies a role in gastrulation. RNA Biology, 9(2): 212-227.
DOI:10.4161/rna.18489 |
Sokol N S, Ambros V. 2005. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Development, 19(19): 2343-2354.
DOI:10.1101/gad.1356105 |
Song Y N, Shi L L, Liu Z Q, Qiu G F. 2014. Global analysis of the ovarian microRNA transcriptome:implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea:Decapoda). BMC Genomics, 15: 547.
DOI:10.1186/1471-2164-15-547 |
Tan T T, Chen M, Harikrishna J A, Khairuddin N, Mohd Shamsudin M I, Zhang G J, Bhassu S. 2013. Deep parallel sequencing reveals conserved and novel miRNAs in gill and hepatopancreas of giant freshwater prawn. Fish & Shellfish Immunology, 35(4): 1061-1069.
DOI:10.1016/j.fsi.2013.06.017 |
Tay Y, Zhang J Q, Thomson A M, Lim B, Rigoutsos I. 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature, 455(7216): 1124-1128.
DOI:10.1038/nature07299 |
Turner L M, Webster S G, Morris S. 2013. Roles of crustacean hyperglycaemic hormone in ionic and metabolic homeostasis in the Christmas Island blue crab, Discoplax celeste. Journal of Experimental Biology, 216(Pt 7): 1191-1201.
DOI:10.1242/jeb.078527 |
Vasudevan S, Tong Y C, Steitz J A. 2007. Switching from repression to activation:microRNAs can up-regulate translation. Science, 318(5858): 1931-1934.
DOI:10.1126/science.1149460 |
Xi Q Y, Xiong Y Y, Wang Y M, Cheng X, Qi Q E, Shu G, Wang S B, Wang L N, Gao P, Zhu X T, Jiang Q Y, Zhang Y L, Liu L. 2015. Genome-wide discovery of novel and conserved microRNAs in white shrimp (Litopenaeus vannamei). Molecular Biology Reports, 42(1): 61-69.
DOI:10.1007/s11033-014-3740-2 |
Xiao M, Li J, Li W, Wang Y, Wu F Z, Xi Y P, Zhang L, Ding C, Luo H B, Li Y, Peng L N, Zhao L P, Peng S L, Xiao Y, Dong S, Cao J, Yu W. 2017. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biology, 14(10): 1326-1334.
DOI:10.1080/15476286.2015.1112487 |
Yan X C, Ding L, Li Y C, Zhang X F, Liang Y, Sun X W, Teng C B. 2012. Identification and profiling of microRNAs from skeletal muscle of the common carp. PLoS One, 7(1): e30925.
DOI:10.1371/journal.pone.0030925 |
Yao X M, Wang L L, Song L S, Zhang H, Dong C H, Zhang Y, Qiu L M, Shi Y H, Zhao J M, Bi Y K. 2010. A Dicer-1 gene from white shrimp Litopenaeus vannamei:expression pattern in the processes of immune response and larval development. Fish & Shellfish Immunology, 29(4): 565-570.
DOI:10.1016/j.fsi.2010.05.016 |
 2018, Vol. 36
2018, Vol. 36