Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CAI Xinglong, CHEN Ying, ZHAO Xiaolu, GAO Jing, AL-FARRAJ Saleh A., YU Lijie, PAN Xuming, QIU Zijian
- Molecular phylogeny and ultrastructure of a new population of Urostyla grandis (Ciliophora, Hypotrichida) from Harbin, China
- Journal of Oceanology and Limnology, 37(1): 256-265
- http://dx.doi.org/10.1007/s00343-019-7277-z
Article History
- Received Oct. 3, 2017
- accepted in principle Dec. 29, 2017
- accepted for publication Jan. 23, 2019
2 Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China;
3 Zoology Department, College of Science, King Saud University, Riyadh 11451, Saudi Arabia
Species of the hypotrich genus Urostyla are frequently distributed in marine, semiterrestrial, and limnetic habitats (Stokes, 1886; Kahl, 1932; Borror, 1979; Alekperov, 1984). Urostyla grandis was originally reported by Ehrenberg (1832), and it was subsequently reported in different names (Ehrenberg, 1832, 1838; Stokes, 1885, 1891; Borror, 1972; Aescht, 2001). Berger(2001, 2006) stated that the type fixation was valid according to the original designation and monotype.
Urostyla species can be distinguished by their body size and outline, size and numbers of the nuclear apparatus, the arrangement and color of the cortical granules, morphology of their oral apparatus, and general somatic ciliary pattern (Stokes, 1886; Kahl, 1932; Suganuma and Inaba, 1967; Ruthmann and Noll-Altmann, 1980; Alekperov, 1984; Song and Wilbert, 1989; Ganner, 1991; Shin, 1994). There were a few reports on the ultrastructure characteristics and identification at the molecular level for Urostyla species (Inaba and Suganuma, 1966; Hoffman and Prescott, 1997; Croft et al., 2003; Hewitt et al., 2003; Dalby and Prescott, 2004; Zhang et al., 2007; Zhao et al., 2009).
Ehrenberg (1838) originally provided the illustration and more or less detailed description of Urostyla grandis, and synonymized Trichoda patens with this species. Later, Stein (1859) provided very detailed living observation of U. grandis, and considered Bursaria vorax Ehrenberg as a junior synonym of U. grandis. Wang (1925) did not illustrate and describe U. grandis in detail. Then Kahl (1932), Reuter (1961), Jerka-Dziadosz (1963), Lundin and West (1963), and Chorik (1968) redescribed U. grandis without discussing them in detail. Jerka-Dziadosz (1972) firstly provided the protargol preparations of U. grandis in which midventral cirri were presented. Jankowski (1979) established the new genus and Metaurostyla polonica based on this population, which Berger (2006) considered to be a junior synonym of U. grandis. Song and Wilber (1989) firstly provided the very detailed morphometric characterization of U. grandis. Croft et al. (2003) made the molecular analyses without supplying any morphologically description in his report.
In this study, a new population of U. grandis was collected from a pound in Harbin, China. Samples were observed in vivo, after protargol silver-staining and by both SEM and TEM. The three-dimensional structure of oral apparatus and the inner surface of the cortex, including microtubules system, blood cell like cortical granules, were illustrated by SEM for the first time. A new type extrusome was found by TEM. In addition, molecular phylogeny was investigated based on small subunit ribosomal DNA (SSU rDNA) sequence data.
2 MATERIAL AND METHOD 2.1 Sample collection and morphological identificationUrostyla grandis was collected on 10 Jun 2016 from a freshwater pond (45°86′N; 126°68′E) in Harbin, Heilongjiang Province, northeastern China (water temperature 20℃ and pH 7.3). Ciliates were maintained and cultured in Petri dishes at 20℃ with Pringsheim solution. Some rice grains were added to promote the growth of bacteria as a food source for the ciliate. Living observation was carried out with bright-field and differential interference contrast microscopy at a magnification of (40–1 250)×. The protargol silver-staining method, following Shi and Frankel (1990), was used to reveal the infraciliature. Measurements of stained specimens were observed at a magnification of 1 250×. Systematics and terminology are mainly according to Lynn (2008).
2.2 Analysis of molecular phylogenyDNA extraction, PCR amplification, and sequencing
Genomic DNA of Urostyla grandis was extracted from purified cells using the DNeasy Tissue Kit (Qiagen, CA) following Gong et al. (2009). The SSU rRNA gene was amplified using universal primers Euk A (5′-AACCTGGTTGATCCTGCCAGT-3′) and Euk B (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) (Medlin et al., 1988). The amplification cycles were as follows: 5 min at 94℃ for initial denaturation; 30 cycles of 94℃ for 30 s (denaturation), 30 cycles of 58℃ for 45 s (annealing), and 30 cycles of 72℃ for 45 s (extension); and a final extension for 8 mins at 72℃. The PCR product was purified using the UNIQ-5 DNA Cleaning Kit (Sangon, Shanghai, China) and inserted into the pMD19-T Simple Vector (TaKaRa, Dalian). Escherichia coli DH 5α cells were used for transformation, and positive clones were identified by PCR screening using the M13-47 and M13-48 primers. Sequencing was performed in both directions on an ABI 3700 sequencer (Invitrogen sequencing facility, Shanghai, China) using primers M13-47, M13-48, as well as two internal primers, namely primer 900F (5′-CGATCAGATACCGTCCTAGT-3′) and 900R (5′-ACTAGGACGGTATCTGATCG-3′).
Phylogenetic analyses
The SSU rRNA gene sequence of our new population of U. grandis and 33 reference sequences from the GenBank database were aligned using the online server GUIDANCE with the alignment algorithm MUSCLE (Penn et al., 2010a, b). The automated removal of unreliable columns was carried out using default parameters (below 0.93), yielding an alignment of 1 720 characters. The sequence identity matrix of five populations of U. grandis were generated using BioEdit 7.0.0 (Hall, 1999).
Four ciliates in the subclasses Oligotrichia and Choreotrichia were chosen as outgroups. Maximum likelihood (ML) analyses were carried out using RAxML-HPC2 on XSEDE v7.2.8 (Stamatakis, 2006; Stamatakis et al., 2008) as implemented in the online CIPRES Science Gateway server (Miller et al., 2010) using the default model. By bootstrapping with 1 000 replicates, the reliability of internal branches was appraised. Bayesian inference (BI) analysis was performed using MrBayes on XSEDE v3.1.2 (Ronquist and Huelsenbeck, 2003) on CIPRES Science Gateway with the GTR+G (0.562 7)+I (0.574 6) model, which was chosen under the Akaike Information Criterion (AIC) in MrModeltest v2 (Nylander, 2004). By using the default settings of a chain length of 1 000 000 generations and trees sampled every 100 generations, Markov Chain Monte Carlo (MCMC) simulations were run with two sets of four chains. The first 25% of sampled trees were discarded as burn-in. By using a majority rule consensus, the posterior probabilities (PPs) were calculated from the remaining trees. MEGA 5 (Kumar et al., 2008) was used to visualize tree topologies.
2.3 Observation of ultrastructureScanning electron microscopy (SEM)
Samples were prepared according to the method described by Gao et al. (2011). Ciliates were fixed in different concentration of KMnO4 for different purposes: 4% KMnO4 for the whole cell preparation; 0.5% KMnO4 for breaking cells preparation. Cells bursts and the cytoplasm overflows by the hypotonic pressure of fixative. After being washed with distilled water for several times, the cytoplasm totally removed, leaving the peeled off pellicle and the exposed inner face. Then the samples were dehydrated, critical-point dried, and gold sprayed by conventional method (Gao et al., 2011). In the last step, ciliates were observed using scanning electron microscopy under 5.00 KV (SEM; HITACHI S-3400).
Transmission electron microscopy (TEM)
According to the method described by Gu et al. (2002), samples were fixed with 2% OsO4 and 5% glutaraldehyde miscible liquids at 4℃ for 30 min. Cells were washed with the sodium cacodylate and gradient acetone dehydration for several times and then embedded with an Epon 812 and polymerized at 40℃ for 20 h and at 60℃ for 48 h. The embedded samples were then ultrathin sectioned and stained with uranyl acetate and lead citrate. At last, they were led for transmission electron microscopy (TEM) observation and photography under 100 KV (FEI TECNAI G2).
3 RESULT 3.1 Morphological identification of live and silver-stained specimens (Fig. 1; Table 1)
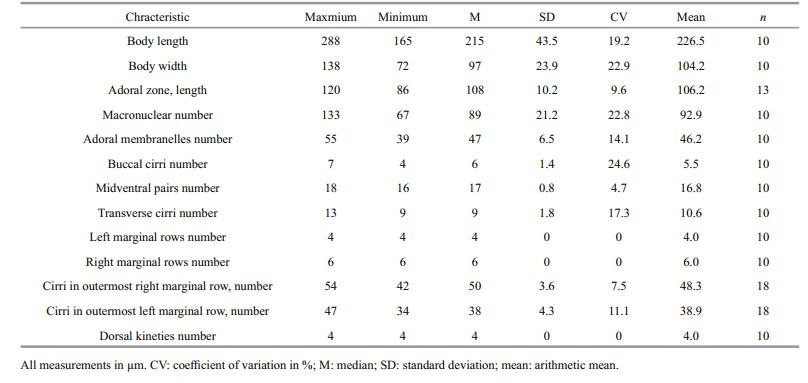
|
| Fig.1 Urostyla grandis in vivo (a–c, e) and after protargol staining (d, f–i) a. ventral view of a representative individual; b. ventral view, to show cortical granules (black arrow) near each cirrus and trichite extrusomes (white arrow); c, d. adoral zone of membranelles (arrows); e. cortical granules arranged in longitudinal rows (arrow); f. dorsal kineties (arrows); g. ventral view, arrows denote midventral complex with cirral pairs in posterior portion of body; h. posterior part of cell, arrows show transverse cirri; i. anterior part, to show buccal cirri (arrows). Scale bars: 60 μm (a, d); 3 μm (b, e); 20 μm (c); 30 μm (f, g, h, i). |
Description of Harbin population: The body size was (165–288)×(72–138) μm in vivo; the body outline was elliptical, with a convex left margin and straight to concave right margin. Both of the anterior and posterior body ends were broadly rounded, while the posterior end curved slightly rightward. About 67–133 macronuclei were found. Contractile vacuoles were found left of the proximal portion of the adoral zone of membranelles (AZM). The cortical granules were about 0.5–1 μm in diameter and colorless, and were found on the ventral side in groups of 2–3 granules near each cirrus (Fig. 1b, black arrow); on the dorsal side, they were arranged in oblique longitudinal rows (Fig. 1e). Somatic cilia about 8 μm long in life. Cytoplasm colourless to slightly grey. Movement slowly sliding.
The AZM occupied one-third of the body length, and comprised of 55–39 membranelles (Fig. 1d). Undulating membranes (UM) were long and curved, which intersected optically slightly below the equator of cell. A row of about three buccal cirri was likely present (Fig. 1i). Slightly enlarged cirri arranged in a bicorona were present. Some smaller cirri were observed between the buccal cirral row and the anterior portion of the midventral complex. The midventral complex comprised cirral pairs (shown in Fig. 1g). Thirteen transverse cirri, arranged in a slightly oblique and subterminal row, are projecting slightly beyond the rear body end (Fig. 1h). About 20 μm in life. Six right marginal rows and four left marginal rows were found in the ventral side. Three dorsal kineties were visible in specimens shown in Fig. 1f. Dorsal bristles about 4 μm long in life.
3.2 Molecular phylogeny based on SSU rRNA gene sequence analyses (Fig. 3; Table 2)
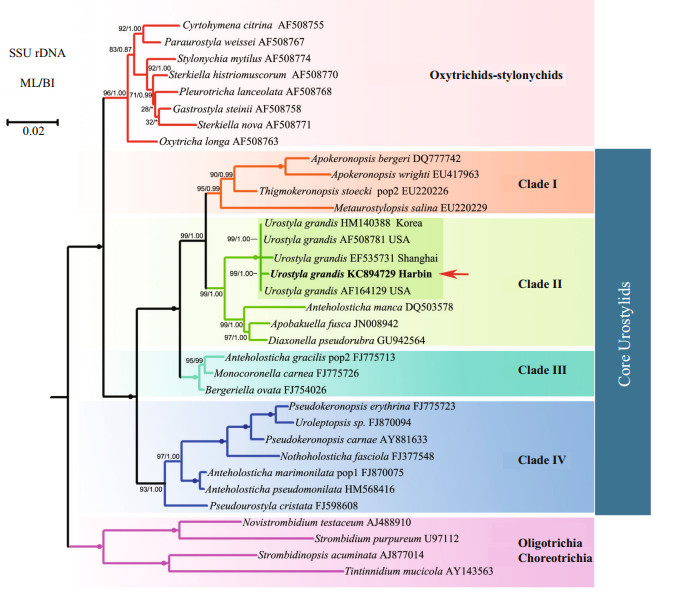
|
| Fig.3 Maximum likelihood (ML) phylogenetic tree based on the small subunit rRNA gene sequences of Urostyla grandis and other spirotrichs. Numbers at the nodes represent the bootstrap values of ML analyses and posterior probability of BI analyses. Fully supported (100%/1.00) branches are marked with solid circles. Asterisk (*) represents disagreement between BI and the reference ML tree. The scale bar corresponds to 2 substitutions per 100 nucleotide positions. Species newly sequenced in the present study is shown in bold and with an arrow. |
|
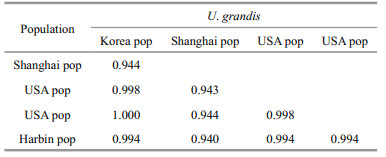
The SSU rRNA gene sequence of the Harbin population of U. grandis was deposited in GenBank with the accession number KC894729. The length and G+C content of the new sequence were 1 767 bp and 44.29%. The SSU rRNA gene sequence similarity of four populations of U. grandis is shown in Table 2. The sequence similarity between the new population found in Harbin, China and other populations ranged from 0.94 to 0.994, which supports the morphological identification of the new population.
The topologies of the SSU rDNA trees constructed using BI and ML analyses were almost identical, therefore only the ML tree is presented here with support values from both algorithms (Fig. 3). Four well-supported, clearly separated clades of core urostylids were recovered in both the BI and ML trees. The Harbin population and the other four populations of U. grandis grouped together in the clade Ⅱ branch of core urostylids with full support (ML 100%, BI 1.00); this is consistent with the morphological classification.
3.3 Ultrastructure descriptions based on SEM and TEM observationsSEM and TEM were combined to study the subpellicular ultrastructure of U. grandis.
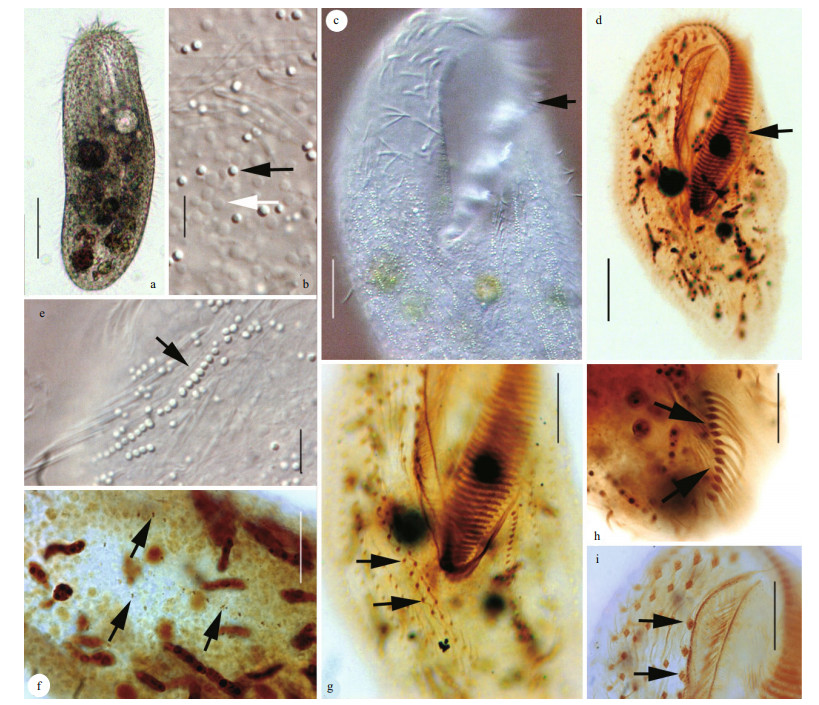
Firstly, the three-dimensional structure of oral apparatus beneath the cortical was illustrated (Fig. 2f). The first microtubule system spread around the root of AZM and separated into two parts (Fig. 2e). The first part, fishnet-like microtubules system, situates on the left of cell along the left margin of the oral apparatus (Fig. 2e, arrows). The second part, paralleled transverse microtubules projecting from the first part, each of which is about 0.6 μm in diameter (Fig. 2e, arrowheads). These paralleled microtubules constructed a rib-like structure for paroral membrane (PM) (Fig. 2l, arrows).
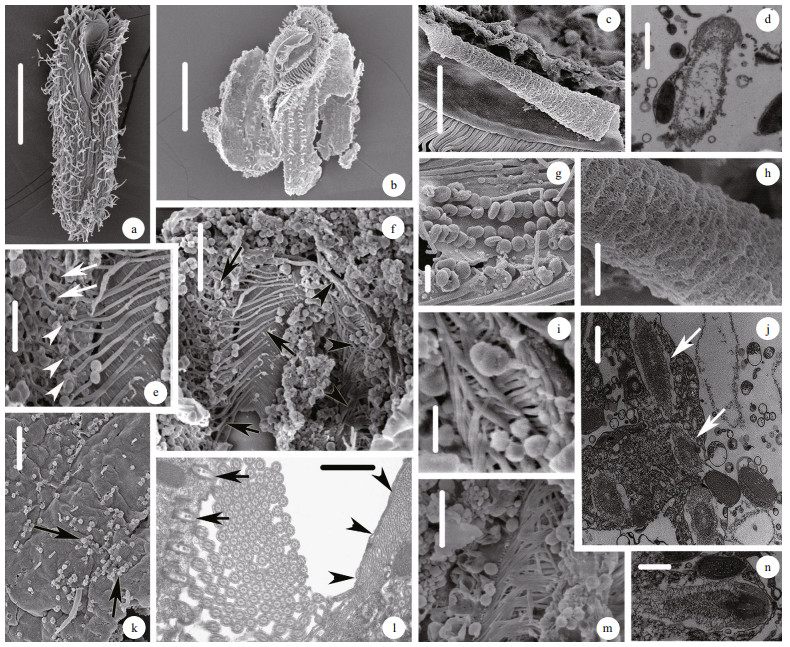
|
| Fig.2 Ultrastructures of Urostyla grandis using both SEM (a-c, e-i, k, m) and TEM (d, j, l, n) a. ventral view of the typical individual; b. external morphology of cell after breaking and washing off the cytoplasm; c. the curly membranelle of oral apparatus; d. mucocyst-like extrusome; e. the first microtubule system. Arrows show the fishnet-like microtubules; arrowheads show the paralleled microtubules; f. the inner microtubule system of oral apparatus. Arrows show the first microtubule system, which contains the fishnet-like microtubules and paralleled microtubules. Arrowheads show the second microtubule system; g. cortical granules appeared as blood cell; h. The inner surface of the membranelle of oral apparatus; i. m. the branches of the second microtubule system; j. cross-section of trichite extrusomes (arrows); k. heads of the extrusomes outside cell surface; l. cross-section of the inner microtubule system of oral apparatus. Arrows show the paralleled microtubules; Arrowheads show the second microtubule system; n. longitudinal-section of trichite extrusomes. Scale bars: 100 μm (a); 50 μm (b); 5 μm (c, e, m); 0.5 μm (d, n); 10 μm (f, k); 1 μm (g, h, j, l); 2.5 μm (i). |
The second microtubule system started from the left top of oral apparatus with three bulky microtubules, each one consisted of bundles of microtubules, which was about 1 μm in diameter (Fig. 2f, arrowheads). Every bulky microtubule forms 3–4 branches after crossing the oral apparatus and extends to the right margin of cell. These branch microtubules interweave together to shape a tree root-like structure (Fig. 2i, m). Above SEM discovery also was supported by TEM (Fig. 2l).
Secondly, two types of extrusomes were found. One is trichite extrusomes and the other is mucocyst-like extrusomes (Fig. 2d, j, k, n). The mucocyst-like extrusomes distributed widely under the pellicle were about 0.5 μm in diameter, long elliptical shape with two parts: the head and the body (Fig. 2d). The trichite extrusomes have three elastic parts: the outerest part was lowest density in three parts and wrapped whole extrusome, the second part situated at the top was the highest density and the third part located at the bottom (Fig. 2j, n). Heads of these extrusomes outside the cell surface were illustrated clearly by SEM (Fig. 2k).
At last, some other structures were also showed by SEM including some blood cell like cortical granules, 0.5–1 μm in diameter (Fig. 2g). The inner surface of the membranelle of oral apparatus with rich microfibers (Fig. 2c, h).
4 DISCUSSION 4.1 Comparison of the Harbin population with other populations of U. grandis in morphological characteristicsSo far, four populations of U. grandis have been well described, as follows: (ⅰ) Song and Wilbert (1989) reported a population from a eutrophic pond in Germany; (ⅱ) Ganner (1991) described two populations: population 1 was from the River Oichten and population 2 was from the Salzach River in Austria; and (ⅲ) Shin (1994) analyzed specimens from limnetic habitats in western South Korea.
It is accepted that the most important criteria for species identification and separation in U. grandis are the body size, the number of adoral membranelles, marginal rows and various cirri (Song and Wilbert, 1989; Ganner, 1991; Shin, 1994). Harbin population can be separated from other population by: 1) body sizes [(165-288)×(72-138) μm in the present work vs. of (162-238)×(72-108) μm in Song and Wilbert (1989) vs. (242.5-330.0)×(92.5-132.5) μm in Ganner (1991) vs. (170-298)×(87-125) μm in Shin (1994)]; 2) The number of macronuclear nodules: [67-133 in the present work vs. 72-142 in Song and Wilbert (1989) vs. 125-227 in Shin (1994)]. 3) The number of adoral membranelles: [39-55 in our study vs. 53-60 in Song and Wilbert (1989) vs. 52-69 in Ganner (1991) vs. 43-64 in Shin (1994)]. 4) The number of buccal cirri: [4-7 in the present work vs. 4-10 (Song and Wilbert, 1989) vs. 7-11 (Ganner, 1991) vs. 6-8 (Shin, 1994)]. 5) The number of transverse cirri: 9–13 in the present work vs. 8–13 in (Song and Wilbert, 1989) vs. 9–15 in (Ganner, 1991) vs. 7–13 in (Shin, 1994)]. 6) Left marginal rows: 4 in present study vs. 6–8 in (Song and Wilbert, 1989) vs. 4–7 in (Ganner, 1991)]. 7) Right marginal rows: [six in the present work vs. 6-8 in (Song and Wilbert, 1989) vs. 5-7 in (Ganner, 1991)]. 8) The number of dorsal kineties: 4 in our study vs. 3–4 in (Song and Wilbert, 1989) vs. 3–4 in (Ganner, 1991) vs. 3 in (Shin, 1994)].
The morphological characters of the Harbin population correspond well with those of populations (ⅰ) to (ⅲ), thus our Harbin population can be identified as U. grandis with no doubt. Nevertheless, we believe these differences to be conspecific.
4.2 SSU rRNA gene sequence analysesThe sequence similarity between the population found in Shanghai, China and other populations ranges from 0.940 to 0.944, which is much lower than those between other populations (around 0.99; Table 2); the phylogenetic tree also shows a relatively long branch of this population (Fig. 3). These may be the result of the direct PCR product sequencing of the Shanghai population, which resulted in errors at the beginning and the ending of the sequence and made a major contribution to the low similarity (Li et al., 2008). The high similarities of SSU rRNA gene sequence show the close relatedness among different populations, and indicates that the SSU rDNA sequence is very stable and conservative.
4.3 Ultrastructure of Urostyla grandisThere are only a few ultrastructural data can be found in several reports of U. grandis populations, e. g. Suganuma and Inaba(1966, 1967), Ruthmann and Noll-Altmann (1980), Bardele (1981), Preisig et al. (1994), Zhang et al. (2007) and Zhao et al. (2009). Inaba and Suganuma (1966) investigated the nuclear apparatus and macronuclear division with electron microscopy. Ruthmann and Noll-Altmann (1980) found that the cytoplasm of U. grandis contains numerous bacteria. The cilia of membranelles and cirri were observed to be covered by a perilemma (Bardele, 1981; Preisig et al., 1994). Zhang et al. (2007) described the mucocysts of U. grandis as investigated using SEM and TEM. Differences of microtubular, macronuclei and cytoplasmic organelles of the ciliate under different physiological status were revealed using TEM by Zhao et al. (2009).
4.3.1 The inner microtubule system of oral apparatusThe cirral patten and oral apparatus of U. grandis were investigated using SEM by Berger (2006). The oral apparatus consists of three parts: adoral zone of membranelles, endoral membranelles and paroral membranelles. A three-dimension structure of the inner microtubule system of oral apparatus and organelles beneath the pellicle had never been reported before. In this paper, a clear inner microtubule system of oral apparatus is reported for the first time, additionally, we found that the inner microtubule system of oral apparatus mentioned above also consists of three parts. This result suggests that the swallow behavior of Urostyla sp. might be controlled by both external and internal structures of the oral apparatus, and external structures of the oral apparatus are tightly connected with the inner microtubule system.
So far, the investigation of the oral apparatus of ciliate only been reported in few species including Paramecium caudatum, Chilodonella cullulus, Tetrahymena vorax, Stylonychida mytilus (Allen and Staehelin, 1981; Smith-Somerville and Buhse, 1984; Sui et al., 2001). Moreover, the oral apparatus of Tetrahymena has been regarded as a useful model for the study of the formation, positioning, and functioning of microtubules and filament systems in ciliated protozoans (Sattler and Staehelin, 1976, 1979; Smith, 1982; Smith-Somerville and Buhse, 1984).
In Tetrahymena vorax, the microtubule system of the oral apparatus can be divided into four parts, namely fine filamentous reticulum (FFR), fibrillary sheets, oral rib region, and cross-connective structure (connection between FFR and fibrillary sheets). As fibrillary sheets collecting the food, the FFR contracted and draw the oral rib walls toward the anterior and left sides. At the same time, the left and anterior portions of the other wall, lined by extensions of oral ribs, would be pulled (Smith-Somerville and Buhse, 1984). According to our new discovered features using SEM and TEM, the structure and mechanism of the oral apparatus in U. grandis are obviously different from those of Tetrahymena species.
4.3.2 The extrusomes and other structuresZhang et al. (2007) have reported that mucocyst was one type of extrusome in U. grandis, which was a long ellipsoid organelle and containing three parts: head, body and tail. In this study, we found that there is another type of extrusome besides the mucocyst. Referred to the description from Rosati and Modeo (2003), we consider the new extrusome to be a type of trichite. In addition, blood cell like cortical granules and the inner surface of membranelle of oral apparatus with microfibers were illustrated for the first time in this paper.
5 CONCLUSIONA ciliate specimen collected from a pound in Harbin were identified and described. The morphology observation by normal taxonomy method and phylogenetic analyses based on SSU rRNA gene sequence supported it was a new population of U. grandis. Moreover, observation on the ultrastructural level provided the complementary data for taxonomic studies of this species, and theoretical support for the possible ingestion mechanism of the ciliates oral apparatus.
6 DATA AVAILABILITY STATEMENTThe original data of SSU rRNA gene sequence of the Harbin population of U. grandis is available from the GenBank under the accession number KC894729. The sequences for all other isolates are available from GenBank with the accession number shown in Fig. 3.
7 ACKNOWLEDGEMENTThe authors would like to express their sincere gratitude to the Deanship of Scientific Research at King Saud University for its funding of this Prolific Research Group (PRG-1436-24). Many thanks are due to Prof. Weibo Song and graduate students at the Laboratory of Protozoology, Ocean University of China, for their technical and institutional help.
Aescht E. 2001. Catalogue of the generic names of ciliates(Protozoa, Ciliophora). Denisia, 1: 1-350.
|
Alekperov I K. 1984. New freshwater ciliates (Hypotrichida)from water bodies of Azerbaijan. Zoologicheskiĭ zhurnal, 63: 1 458-1 463.
|
Allen R D, Staehelin L A. 1981. Digestive system membranes:freeze-fracture evidence for differentiation and flow in Paramecium. The Journal of Cell Biology, 89(1): 9-20.
|
Bardele C F. 1981. Functional and phylogenetic aspects of the ciliary membrane:a comparative freeze-fracture study. Biosystems, 14(3-4): 403-421.
DOI:10.1016/0303-2647(81)90046-0 |
Berger H. 2001. Catalogue of Ciliate Names 1. Hypotrichs.Verlag Helmut Berger, Salzburg. 206p.
|
Berger H. 2006. Monograph of the Urostyloidea (Ciliophora, Hypotricha). Springer, Dordrecht. p.1-1 303.
|
Borror A C. 1972. Revision of the order Hypotrichida(Ciliophora, Protozoa). The Journal of Protozoology, 19(1): 1-23.
DOI:10.1111/j.1550-7408.1972.tb03407.x |
Borror A C. 1979. Redefinition of the Urostylidae (Ciliophora, Hypotrichida) on the basis of morphogenetic characters. The Journal of Protozoology, 26(4): 544-550.
DOI:10.1111/j.1550-7408.1979.tb04192.x |
Chorik F P. 1968. Free-living Ciliates in Moldavian Water Basins. Akadmia Nauk MSSR, Kischinev. 251p. (in Russian)
|
Croft K E, Dalby A B, Hogan D J, Orr K E, Hewitt E A, Africa R J, DuBois M L, Prescott D M. 2003. Macronuclear molecules encoding actins in spirotrichs. Journal of Molecular Evolution, 56(3): 341-350.
DOI:10.1007/s00239-002-2405-2 |
Dalby A B, Prescott D M. 2004. The scrambled actin Ⅰ gene in Uroleptus pisces. Chromosoma, 112(5): 247-254.
DOI:10.1007/s00412-003-0270-4 |
Ehrenberg C G. 1832a. Beiträge zur kenntniss der organisation der infusorien und ihrer geographischen verbreitung, besonders in Sibirien. Abhandlungen der Preussischen Akademie der Wissenschaften, 1830: 1-88.
|
Ehrenberg C G. 1832b. Űber die entwickelung und lebensdauer der infusionsthiere; nebst ferneren Beiträgen zu einer Vergleichung ihrer organischen Systeme. Abhandlungen der Königlichen Akademie Wissenschaften zu Berlin, Physikalische Klasse, 1831: 1-154.
|
Ehrenberg C G. 1838. Die Infusionsthierchen als Vollkommene Organismen-Ein Blick in das Tiefere Organische Leben der Natur. Leopold Voss, Leipzig. 548p.
|
Ganner B. 1991. Zur Taxonomie einiger Ciliaten (Protozoa)aus Fliessgewässern. Dissertation Universität Salzburg. 159p.
|
Gao J, Zhou W, Qiao L, Chen Y, Qiu Z J. 2011. A new method of sample preparation for scanning electron microscope used to observe the structure under pellicle in ciliates. Chinese Journal of Cell Biology, 33(9): 1 004-1 007.
(in Chinese with English abstract) |
Gong J, Stoeck T, Yi Z Z, Miao M, Zhang Q Q, Roberts D M, Warren A, Song W B. 2009. Small subunit rRNA phylogenies show that the class Nassophorea is not monophyletic (Phylum Ciliophora). Journal of Eukaryotic Microbiology, 56(4): 339-347.
DOI:10.1111/j.1550-7408.2009.00413.x |
Gu F K, Chen L, Ni B, Zhang X M. 2002. A comparative study on the electron microscopic enzymo-cytochemistry of Paramecium bursaria from light and dark cultures. European Journal of Protistology, 38(3): 267-278.
DOI:10.1078/0932-4739-00875 |
Hall T A. 1999. BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95-98.
|
Hewitt E A, Müller K M, Cannone J, Hogan D J, Gutell R, Prescott D M. 2003. Phylogenetic relationships among 28 spirotrichous ciliates documented by rDNA. Molecular Phylogenetics and Evolution, 29(2): 258-267.
DOI:10.1016/S1055-7903(03)00097-6 |
Hoffman D C, Prescott D M. 1997. Phylogenetic relationships among hypotrichous ciliates determined with the macronuclear gene encoding the large, catalytic subunit of DNA polymerase α. Journal of Molecular Evolution, 45(3): 301-310.
DOI:10.1007/PL00006234 |
Inaba F, Suganuma Y. 1966. Electron microscopy of the nuclear apparatus of urostyla grandis, a Hypotrichous ciliate. The Journal of Protozoology, 13(1): 137-143.
DOI:10.1111/j.1550-7408.1966.tb01884.x |
Jankowski A W. 1979. Revision of the order Hypotrichida Stein, 1859. Generic catalogue, phylogeny, taxonomy.Academy of Science USSR, Proceedings of the Zoological Institute, 86: 48-85.
|
Jerka-Dziadosz M. 1963. Morphogenesis in division and regeneration of Urostyla grandis Ehrbg. Acta Protozool, 1: 43-54.
|
Jerka-Dziadosz M. 1972. Cortical development in Urostyla. Ⅰ.comparative study on morphogenesis in U. cristata and U.grandis. Acta Protozool, 10: 73-100.
|
Kahl A. 1932. Urtiere oder Protozoa Ⅰ:Wimpertiere oder Ciliata (Infusoria) 3. Spirotricha. Tierwelt Dtl, 25: 399-650.
|
Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA:a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics, 9(4): 299-306.
DOI:10.1093/bib/bbn017 |
Li Y S, Niu Y N, Liu L X. 2008. Phylogenetic studies of four species of ciliate inferred from 16s-like small subunit rRNA gene sequences. Journal of Forestry Research, 19(2): 119-124.
DOI:10.1007/s11676-008-0020-9 |
Lundin F C, West L S. 1963. The Free-living Protozoa of the Upper Peninsula of Michigan ([Northern Michigan College] Monographic series). Northern Michigan College Press, Marquette. 175p.
|
Lynn D H. 2008. The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature. 3rd edn.Springer, Dordrecht. p.1-605.
|
Medlin L, Elwood H J, Stickel S, Sogin M L. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene, 71(2): 491-499.
DOI:10.1016/0378-1119(88)90066-2 |
Miller M A, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of 2010 Gateway Computing Environments Workshop. IEEE, New Orleans, LA. p.1-8, https://doi.org/10.1109/GCE.2010.5676129.
|
Nylander J A A. 2004. MrModeltest Version 2. Program Distributed by the Author. Uppsala University, Uppsala.
|
Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. 2010b. GUIDANCE:a web server for assessing alignment confidence scores. Nucleic Acids Research, 38(S2): W23-W28.
DOI:10.1093/nar/gkq443 |
Penn O, Privman E, Landan G, Graur D, Pupko T. 2010a. An alignment confidence score capturing robustness to guide tree uncertainty. Molecular Biology and Evolution, 27(8): 1 759-1 767.
DOI:10.1093/molbev/msq066 |
Preisig H R, Anderson O R, Corliss J O, Moestrup Ø, Powell M J, Roberson R W, Wetherbee R. 1994. Terminology and nomenclature of protist cell surface structures. Protoplasma, 181(1-4): 1-28.
DOI:10.1007/BF01666386 |
Reuter J. 1961. Einige faunistische und ökologische Beobachtungen über Felsentümpel-Ziliaten. Acta zoologica Fennica, 99: 1-42.
|
Ronquist F, Huelsenbeck J P. 2003. MRBAYES 3:Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1 572-1 574.
DOI:10.1093/bioinformatics/btg180 |
Rosati G, Modeo L. 2003. Extrusomes in ciliates:diversification, distribution, and phylogenetic implications. The Journal of Eukaryotic Microbiology, 50(6): 383-402.
DOI:10.1111/j.1550-7408.2003.tb00260.x |
Ruthmann A, Noll-Altmann G. 1980. Effect of bacterial symbiont on cell division in a ciliate. In: Schwemmler W, Schenk H E A eds. Endocytobiology: Endosymbiosis and Cell Biology Volume 1. Walter de Gruyter & Co, Berlin, New York. p.361-370.
|
Sattler C A, Staehelin L A. 1976. Reconstruction of oral cavity of Tetrahymena pyriformis utilizing high voltage electron microscopy. Tissue and Cell, 8(1): 1-18.
DOI:10.1016/0040-8166(76)90016-1 |
Sattler C A, Staehelin L A. 1979. Oral cavity of Tetrahymena pyriformis:a freeze-fracture and high-voltage electron microscopy study of the oral ribs, cytostome, and forming food vacuole. Journal of Ultrastructure Research, 66(2): 132-150.
DOI:10.1016/S0022-5320(79)90130-8 |
Shi X B, Frankel J. 1990. Morphology and development of Mirror-Image doublets of Stylonychia mytilus. The Journal of Protozoology, 37(1): 1-13.
DOI:10.1111/j.1550-7408.1990.tb01106.x |
Shin M K. 1994. Systematics of Korean Hypotrichs(Ciliophora, Polyhymenophora, Hypotrichida) and Molecular Evolution of Hypotrichs. Seoul National University, Seoul. 270p.
|
Smith H E. 1982. Oral apparatus structure in the carnivorous macrostomal form of Tetrahymena vorax. The Journal of Protozoology, 29(4): 616-627.
DOI:10.1111/j.1550-7408.1982.tb01348.x |
Smith-Somerville H E, Buhse Jr H E. 1984. Changes in oral apparatus structure accompanying vacuolar formation in the macrostomal form of Tetrahymena vorax. A model for the formation of food vacuoles in Tetrahymena. The Journal of Protozoology, 31(3): 373-380.
DOI:10.1111/j.1550-7408.1984.tb02982.x |
Song W B, Wilbert N. 1989. Taxonomische untersuchungen an aufwuchsciliaten (protozoa, ciliophora) im poppelsdorfer weiher, Bonn. Lauterbornia, Heft, 3: 2-221.
|
Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology, 57(5): 758-771.
DOI:10.1080/10635150802429642 |
Stamatakis A. 2006. RAxML-Ⅵ-HPC:maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22(21): 2 688-2 690.
DOI:10.1093/bioinformatics/btl446 |
Stein F. 1859. Der Organismus der Infusionsthiere Nach Eigenen Forschungen in Systematischer Reihenfolge Bearbeitet. Ⅰ. Engelmann, Leipzig. 206p.
|
Stokes A C. 1885. XL.-Some new infusoria from American fresh waters. Annals and Magazine of Natural History:Series 5, 15(90): 437-449.
DOI:10.1080/00222938509459365 |
Stokes A C. 1886. Some new Hypotrichous infusoria. Proceedings of the American Philosophical Society, 23(121): 21-30.
|
Stokes A C. 1891. Notes of new infusoria from the fresh waters of the United States. Journal of the Royal Microscopical Society, 11(6): 697-704.
DOI:10.1111/j.1365-2818.1891.tb01611.x |
Suganuma Y, Inaba F. 1966. Electron microscopy of the macronucleus in interphase of Urostyla grandis. Doubutsugaku Zasshi, 75(6): 172-177.
|
Suganuma Y, Inaba F. 1967. Electron microscopic study of the macronuclei of Urostyla grandis during binary fission. Zoological magazine, 76(4): 110-118.
|
Sui S G, Chen Y, Qiu Z J, Shi X B. 2001. Structure of oral apparatus in Stylonychia mytilus. Acta Zoologica Sinica, 47(4): 436-441.
(in Chinese with English abstract) |
Wang C C. 1925. Study of the protozoa of Nanking Part Ⅰ.Contr. Boil. Lab. Sci. Soc. China, 1: 1-60.
|
Zhang J, Ni B, Sheng C, Gu F K. 2007. Observations on the ultrastructure of mucocyst in Urostyla grandis. Journal of Fudan University (Natural Science), 46(6): 972-975.
(in Chinese with English abstract) |
Zhao L, Li Y S, Li J G, Gu F K. 2009. Some ultrastructural observations of the vegetative, resting and excysting ciliate, urostyla grandis (urostylidae, hypotrichida). Biological Research, 42(4): 395-401.
DOI:10.4067/S0716-97602009000400001 |
 2019, Vol. 37
2019, Vol. 37