Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Shuping, CHEN Fangyi, ZHANG Yaqun, MA Xiaowan, QIAO Kun
- Gap junction gene innexin3 being highly expressed in the nervous system and embryonic stage of the mud crab Scylla paramamosain
- Journal of Oceanology and Limnology, 37(5): 1649-1658
- http://dx.doi.org/10.1007/s00343-019-8096-y
Article History
- Received Apr. 17, 2018
- accepted in principle Aug. 28, 2018
- accepted for publication Dec. 19, 2019
2 State-Province Joint Engineering Laboratory of Marine Bioproducts and Technology, Xiamen University, Xiamen 361005, China;
3 Fujian Collaborative Innovation Center for Exploitation and Utilization of Marine Biological Resources, Xiamen University, Xiamen 361005, China;
4 State Key Laboratory of Environmental Criteria and Risk Assessment, Chinese Research Academy of Environmental Sciences, Beijing 100012, China
Gap junctions in vertebrates enable the transfer of ions and small signal molecules (< 1 kD) including second messengers directly between adjacent cells and play a vital role in development, neural synchronization, signaling, muscular contraction, metabolic cooperation, exocrine secretion and homeostasis (Nagy et al., 2004; McCracken and Roberts, 2006; Kielian, 2008; Bosco et al., 2011). Gap junctions are formed of homomeric or heteromeric half-junctions docking on two adjacent cells. It has been shown that several innexin proteins oligomerize to form a half-channel in each cell and two halfchannel docks to form a gap junction channel (Altun et al., 2009; Abascal and Zardoya, 2013; Oshima et al., 2013, 2016). Innexins have several features in common with their vertebrate counterparts named connexins but lack a primary amino acid sequence homology (Phelan et al., 1998; Skerrett and Williams, 2017).
Innexin genes have been found in different invertebrates, such as Drosophila melanogaster, Clione limacine, Schistocerca Americana, Giardia tigrin, Chaetopterus variopedatus, Hirudo medicinalis, Hydra vulgaris, Caenorhabditis elegans, and Aedes aegypti (Bauer et al., 2002; Phelan, 2005; Altun et al., 2009; Abascal and Zardoya, 2013; Calkins et al., 2015). Eight innexin genes are expressed in D. melanogaster (Stebbings et al., 2002), 25 innexin genes in C. elegans (Altun et al., 2009), 21 innexin genes in H. verbena (Kandarian et al., 2012), 6 innexin genes in Cancer borealis (Shruti et al., 2014) and 6 innexin genes in Homarus americanus (Northcutt et al., 2016).
Innexins have been previously studied in epithelia tissues, foregut, ovary, testis, brain, and stomatogastric ganglion (Bauer et al., 2004; Dykes and Macagno, 2006; Bohrmann and Zimmermann, 2008; Hong et al., 2008; Northcutt et al., 2016). Gap junctions which are composed of innexin proteins in the nervous system are considered as components of electrical synapses and may play an important role in the physiological regulation of the nervous system (Phelan et al., 1998; Phelan et al., 2008; Wu et al., 2011). Researches have shown that the gap junctions between optic fibers and target optic lamina neurons can form synaptic circuitry in Daphnia (Lopresti et al., 1974). The expression of the innexin genes shakB and ogre in the visual system during Drosophila pupal development might play a role in the development of normal chemical synapses between laminar neurons and photoreceptors (Curtin et al., 2002).
Innexin genes have been demonstrated to play vital roles in embryonic development and morphogenesis in invertebrates. Innexin3 gene of C. elegans is highly expressed in the embryogenesis period but weakly expressed at the postembryonic period (Starich et al., 2003). The loss of innexin3 led to dorsal closure defects in Drosophila embryo (Giuliani et al., 2013). Lechner et al. (2007) suggested that the transcriptional cross regulation of paracrine and gap junctionsmediated signaling is essential for organogenesis in Drosophila. The innexin2 gene of D. melanogaster is the target gene of the conserved WNT signal pathway that controls the morphosis of the epithelial tissues and organs (Bauer et al., 2002).
Revealing the expression and characteristics of innexins in the embryonic nervous system is imperative for understanding development and is a rapidly expanding field of developmental neurobiology. Innexin1 gene of S. americana mainly expresses in the embryonic nervous system, especially the neural precursor cells, and neurogliocyte, while the innexin2 gene predominantly expresses in the embryonic neural precursor cells (Ganfornina et al., 1999). The innexin gene ogre mutation in Drosophila causes a decreased number of neurons in the optic lobes and ogre is required in glial cells for normal postembryonic development of the central nervous system, which indicates the role of this gene in neurogenesis (Watanabe and Kankel, 1992; Holcroft et al., 2013). The innexin gene nsy-5 interacts with a calcium channel-CaMKII kinase pathway, and they co-regulate gene expression patterns, synaptic protein distribution, and sensory specificity in the left and right AWC neurons (Chuang et al., 2007).
Scylla paramamosain is an important commercial breeding crab in the coastal areas of South Eastern China. Until now, only one innexin gene named Sp-inx2 has been found by our laboratory. Sp-inx2 could form hemichannels in the crab hemocytes and regulate immune response and cell apoptosis of S. paramamosain (Wang et al., 2015). However, the existence and function of other innexin family members in S. paramamosain have not been studied yet. In this study, we cloned the full length of the cDNA sequence of Sp-inx3 gene by RACE PCR and analyzed its tissue distribution through qPCR and western blotting methods. Then, brain and thoracic ganglion mass were selected for IHC assay. Furthermore, we investigated the Sp-inx3 transcription profile at different development stages of the crab. This work will provide insight into the role of Sp-inx3 in early embryonic development and the nervous system of S. paramamosain.
2 MATERIAL AND METHOD 2.1 Experimental animals and sample collectionLive healthy male and female S. paramamosain were purchased from a commercial crab farm in Xiamen, China. The average body weight of these crabs was 300±45 g.
Hemocytes were separated from the haemolymph using the method described previously (Wang et al., 2015). Tissues and organs including eyestalk (EY), brain (BR), gills (GI), ovaries (OA), heart (HT), muscle (MU), stomach (ST), spermathecae (SP), hepatopancreas (HP), midgut gland (MG), reproductive tract (RT), thoracic ganglion mass (TG) from female crabs and EY, BR, TG, HC from male crabs were sampled individually (n=5), and immediately frozen in liquid nitrogen, and stored at -80℃ to be used later. Early to late-stage embryos including embryo 1–4, prehatching and the zoea 1 larva stage of S. paramamosain were collected, as shown in our previous study (Chen et al., 2015).
2.2 Cloning of Sp-inx3 full length cDNA sequenceAccording to the nucleotide sequence of Cancer borealis innexin3 gene (GenBank, JQ994481.1), primers Sp-inx3F (5′-ATGATGAAATATTTGGCAGCCGCG-3′) and Sp-inx3R (5′-CAACATTGGTTTCCGTTCCAGG-3′) were used to obtain the partial sequence of Sp-inx3. Primers Sp-inx1F (5′-GTGTTGATCGGCGCCTCTGTCCT-3′) and Sp-inx1R (5′-TACAGACCCGCACTGTTGTA-3′) were also used to obtain the partial sequence of Sp-inx1 according to the Cancer borealis innexin1 gene (GenBank, JQ994479.1). The resulting fragment was then cloned into a pMD-18T vector (TaKaRa, Japan) and sequenced (Sangon Biotech, Shanghai, China). Based on the partial cDNA sequence of Sp-inx3, RACE PCR was employed to obtain its full-length cDNA sequence by using primers Sp-inx3 5′ (5′-CACTATCTCGTCATCAGGCTCGGCATGG-3′) and Sp-inx3 3′ (5′-AGCTGGAGAATTCAGTTTCAACCCTG-3′). RACE PCR was also employed to amplify the full-length cDNA of Sp-inx1 gene by using primers Sp-inx1 5′ (5′-CGACCCACTGGTAGTAGTTGTGGAAGCG-3′) and Sp-inx1 3′ (5′- CGAGACTGCCAACAGCAACAGCCCC-3′). The RACE PCR was performed according to the manufacturer's instructions of the RACE Core Set (TaKaRa, Japan).
The ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to predict amino acid sequences of Sp-inx3. Sp-inx1, Sp-inx2, Sp-inx3 protein sequence, and innexin protein family from D. melanogaster and C. elegans were used for multiple sequence alignment. An evolutionary tree of innexins was constructed by the neighbor-joining method with MEGA software, version 6.0.
2.3 Determination of Sp-inx3 mRNA expression profile by qPCRRNA was extracted from different individual samples using Trizol reagent (RNA Extraction Kit, Invitrogen, Carlsbad, CA, USA) and the first-strand cDNA were synthesized with PrimeScript RT reagent kit (Perfect Real Time) (Takara, Japan) following the manufacturer's instruction. qPCR was carried out as described in our previous study (Wang et al., 2015). Amplifications were carried out in a 20-μL reaction tube containing 10 μL Power SYBR Green PCR Master Mix (Applied Biosystems, UK, ABI) and 5 ng of total transcribed cDNA. The procedure was: 50℃ for 2 min, 95℃ for 10 min, 40 cycles of 95℃ for 15 s, and 60℃ for 1 min. Amplifications were repeated in triplicate. The expression level of Sp-inx3 was analyzed by the 2-ΔΔCt method (Livak and Schmittgen, 2001). β-actin gene was employed as the reference gene. The primers qF (5′-ACGAGGCACAAGAAGCA-3′) and qR (5′-GGAACAGGTCGGTGAAGTA-3′) were used for Sp-inx3 gene amplification. Primers β-actin F (5′-GCCCTTCCTCACGCTATCCT-3′) and β-actin R (5′-GCGGCAGTGGTCATCTCCT-3′) were used for β-actin gene amplification.
2.4 Western blotting and IHC assayThe sequence located in COOH-terminal domain of Sp-inx3 (NH2-KKCNLGDWFLIYHLGRNMEPMV YGEFLKEFAKELENSVSTLERKPMLVGS-COOH) was amplified using the primers: Sp-inx3 CTF (5′-GCGCCATGGGCAAGAAGTGCAACCTTGGAGACTGGT-3′) and Sp-inx3 CTR (5′-GGGCTCGAGACTTCCTACCAACATTGGTTTAC-GT-3′). The PCR product was inserted into the pET-28a(+) vector (Novagen, San Diego, CA, USA). The restriction enzyme recognition sites were Xho I and Nco I. The protocol of plasmid construction and expression of recombinant protein was similar to that described previously (Wang et al., 2015). The preparation process of the polyclonal antibody against Sp-inx3 was the same as that for Sp-inx2 (Wang et al., 2015).
The various tissues and developmental stage samples were lysed by RIPA lysate. Protein concentration was measured by the Pierce BCA protein assay (Thermo Scientific, Rockford, IL, USA). Protein extractions were separated by SDSPAGE and transferred to a PVDF membrane (BIORAD, Hercules, CA, USA) using wet blotting. Membranes were blocked in 5% skin non-fat milk and 0.3% Tween 20 in PBS, probed with primary antibodies, and goat anti-mouse IgG HRP-conjugated antibody was used as the secondary antibody. Positive signals of individual blots were detected using enhanced chemiluminescence Western blotting kit (Millipore, Billerica, MA, USA).
The brain and thoracic ganglion mass of female crabs were sampled and fixed with 4% paraformaldehyde overnight and dehydrated in different ethanol series. After clearing with xylene, they were embedded in paraffin and cut into 6–7 μm thickness. After the deparaffinization and rehydration steps, the sections were heated with citrate buffer (0.01 mol/L, pH 6.0) for 3 min. For blocking the endogenous peroxidase activity, these sections were incubated in 3% hydrogen peroxide in methyl alcohol for 20 min. To block non-specific protein-protein interactions, the sections were incubated with the blocking buffer (1% BSA/0.3 mol/L glycine/10% normal goat serum (Solarbio, Beijing, China) in 0.1% PBS-Tween) for 1 h. Then the sections were incubated with mouse polyclonal anti-Sp-inx3 antibody (1:1 000 dilutions) for 1 h and pre-immune serum was the control. Goat anti-mouse IgG (1:1 000 dilutions) was used as the secondary antibody to incubate with these sections for 45 min. DAB solution (Maixin Biotechnology, Fuzhou, China) was used to stain the positive signal. All the sections were examined by an inverted microscope (Leica, Heidelberg, Germany).
2.5 Statistical analysisData were presented as means ± SD. Statistical analysis was performed using SPSS 16.0 software (SPSS Inc, Chicago, USA). One-way ANOVA was conducted to analyze multiple group comparison. Statistical differences were judged to be significant when P < 0.05.
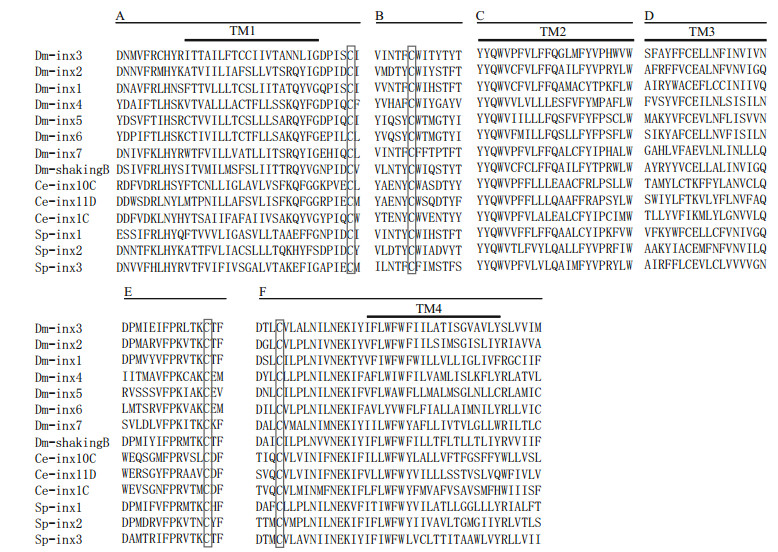
3 RESULT 3.1 Characterization of Sp-inx3 cDNA and protein sequenceThe cloned full-length cDNA sequence of Sp-inx3 is 1 869 bp (GenBank, MG324349), with a 64-bp 5′-untranslated region (UTR), a 701-bp 3′-UTR, and an open reading frame (ORF) of 1 101 bp encoding 367 amino acids. The deduced molecular weight was 42.3 kDa with an isoelectric point (pI) of 9.10 (Fig. 1). The most conserved parts of this protein family include the extracellular loops, the "YYQWV" motif and one proline and one tryptophan near the end of the second transmembrane domain (Abascal and Zardoya, 2013; Wang et al., 2015) (Fig. 2).

|
| Fig.1 The complete cDNA sequence of Sp-inx3 and its deduced amino acid sequence (below) * represents termination codon; gray shadows show deduced transmembrane domains; the circles represent the cysteine residues in the extracellular loops. The bold "ATG" represents initiation codon, the bold "TAA" represents termination codon, and the bold sequence "ATTTAAA"is the polyadenylation signal sequence in the 3' non-coding region. |
|
| Fig.2 Multiple sequences alignment of Sp-inx3 protein with innexin protein family Only highly conserved regions appeared in the figure. The innexin proteins which were downloaded from GenBank including: Dm-inx1 (P27716), Dminx2 (AAD50379), Dm-inx3 (AAF50922), Dm-inx4 (AAF48923), Dm-inx5 (AAL25820), Dm-inx6 (AAL25821), Dm-inx7(Q9V3W6), Dm-shakingB (NP_728361.1), Ce-inx1C (AGV53074.1), Ce-inx10C (AGV53075.1), Ce-inx11D (AGV53078.1), Sp-inx1 (MG324350), Sp-inx2 (FJ774668.1), Sp-inx3 (MG324349). A. first transmembrane domains and the N-terminal and C-terminal flanking regions; B. conserved regions in the first extracellular loops; C. second transmembrane domains and the conserved peptide YYQWV at the end of the first extracellular loops; D. third transmembrane domains and the C-terminal flanking regions; E. conserved regions in the second extracellular loops; F. part of the fourth transmembrane domains and the conserved region in the second extracellular loops. |
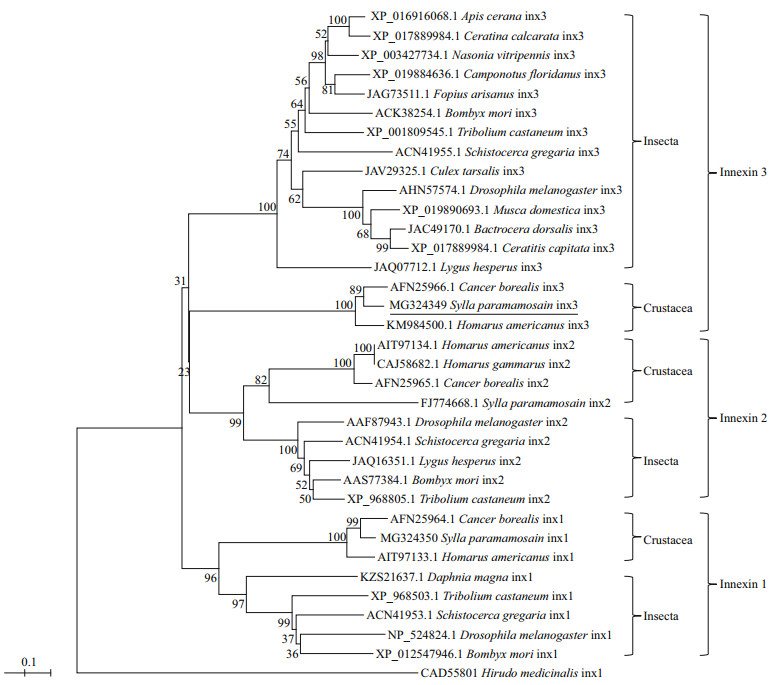
To study the phylogenetic relationship of Sp-inx3 protein with other known innexins, an evolutionary tree was constructed using the neighbor-joining method (Fig. 3). The result shows two major clades within the innexin family. The first clade was divided into three branches: the innexin3 proteins in insects or in crustaceans and all of the innexin2 proteins. The second clade included all the innexin1 proteins. The sp-inx3 protein showed the highest amino acid identity (88.0%) to Cancer boteals innexin3 and had a relatively high identity (83%) with Homarus americanus innexin3.
|
| Fig.3 Phylogenetic analysis of Sp-inx3 with other known innexins The phylogenetic tree based on amino acid sequences was constructed by the neighbor-joining method with MEGA5. The GenBank accession numbers of innexins are also listed in the figure. |
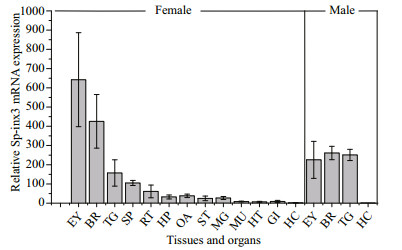
The qRT-PCR results showed that Sp-inx3 gene is expressed predominantly in the nervous system of both female and male crabs, with the highest level of transcripts in the eyestalk, followed by brain and thoracic ganglion mass in female crabs (Fig. 4). Sp-inx3 gene transcription could also be detected in other tissues including spermatheca, reproductive tract, hepatopancreas, ovary, stomach, midgut gland, muscle, heart, and gills in female crabs, but at a much lower level. One exception is the hemocytes, in which the expression of Sp-inx3 mRNA was not detectable in either female or male crabs.
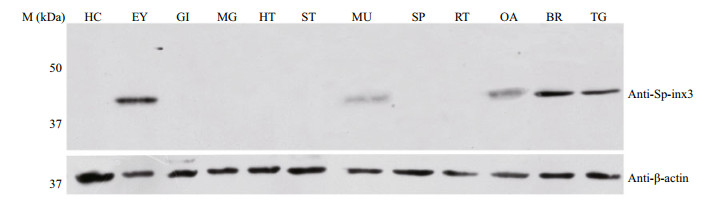
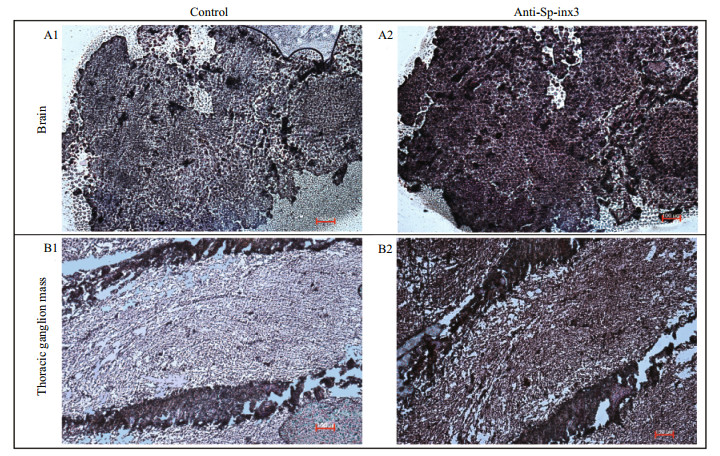
Like the transcription pattern of Sp-inx3 in various tissues, the Sp-inx3 protein was mainly expressed in the eyestalk, brain and thoracic ganglion mass (Fig. 5). Brain and thoracic ganglion mass were chosen for IHC assay. The result also shows the widespread and intense immunoreactivity of Sp-inx3 in both tissues and no positive signal was detected in the negative mouse serum treatment groups (Fig. 6).
|
| Fig.4 Expression levels of Sp-inx3 mRNA in different tissues measured by qRT-PCR RNA samples were extracted from EY: eyestalk; BR: brain; TG: thoracic ganglion mass; SP: Spermatheca; RT: reproductive tract; HP: hepatopancreas; OA: ovary; ST: stomach; MG: midgut gland; MU: muscle; HT: heart; GI: gills; HC: hemocytes. |
|
| Fig.5 Sp-inx3 protein expression in tissues and organs of female S. paramamosain Crude protein lysates samples were extracted from HC: hemocytes; EY: eyestalk; GI: gills; MG: midgut gland; HT: heart; ST: stomach; MU: muscle; SP: Spermatheca; RT: reproductive tract; OA: ovary; BR: brain; TG: thoracic ganglion mass. |
|
| Fig.6 Distribution of Sp-inx3 protein in the brain and thoracic ganglion mass of S. paramamosain by IHC A1 & B1: the brain and thoracic ganglion mass sections incubated with pre-immune serum; A2&B2: the brain and thoracic ganglion mass sections incubated with mouse polycolonal anti-Sp-inx3 antibody (1:1 000 dilutions). Scale bars represent 100 μm. |
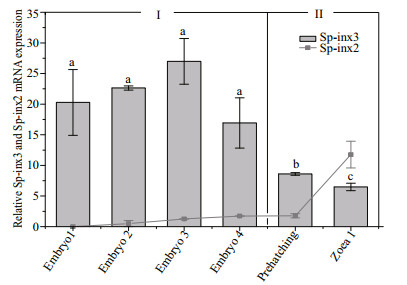
The transcripts of Sp-inx3 from early to late embryos stage including embryo 1-4, prehatching and the zoea 1 larvae stage of S. paramamosain were examined. As shown in Fig. 7, Sp-inx3 gene maintained a high level of transcription from the embryo1 to embryo4 period, then gradually reduced from the pre-hatching period and fell to the lowest level at the zoea 1 larvae stage. There were no significant differences in the mRNA level in embryo1, embryo2, embryo3 and embryo4 periods (P>0.05). The Sp-inx3 mRNA level at the pre-hatching period was 32% of the embryo3 period (P < 0.05) and its level at the zoea larvae stage 1 was 24% of the embryo3 period (P < 0.05). In summary, the Sp-inx3 transcription profile in the embryo could be divided into two stages: (Ⅰ) the period before pre-hatching and (Ⅱ) prehatching period and zoea 1 larvae. Figure 7 also shows that Sp-inx2 mRNA was not detected in the embryo1 period and its transcription level gradually increased from the embryo1 period to the zoea larvae stage Ⅰ. Moreover, the transcription level of Sp-inx3 was significantly higher than that of Sp-inx2 from the embryo1 period to the pre-hatching period (P < 0.05) while it was lower in the zoea larvae 1.
|
| Fig.7 Sp-inx3 and Sp-inx2 gene expression during different developmental stages of S. paramamosain |
Despite S. paramamosain being an important commercial species, its important role in understanding arthropod evolution and as a model for neurobiology and developmental biology have been overlooked. Previous studies revealed that innexins might be developmentally regulated to express and play crucial roles in the operation of neuronal networks (Ducret et al., 2006; Chuang et al., 2007; Holcroft et al., 2013). Therefore, it is interesting to reveal the role of Sp-inx3 in the nervous system and developmental stages of S. paramamosain. The key finding of this study is that Sp-inx3 gene was highly expressed in the nervous system including the eyestalk, brain and thoracic ganglion mass, and Sp-inx3 mRNA expression in embryonic development was divided into two distinct phases with a higher transcription level in early embryonic development.
We have identified three innexin family members: Sp-inx1, Sp-inx2, and Sp-inx3. They contain similar amino acid residue lengths (380 aa, 359 aa, and 360 aa, respectively). However, their isoelectric points are different (6.46 for Sp-inx1, 8.63 for Sp-inx2, and 9.10 for Sp-inx3). The most conserved regions of the proteins are the four transmembrane sequences and the YYQWV motif was the signature sequence in all innexin proteins (Yen and Saier, 2007).
In addition, Sp-inx3 protein has similar protein structure characteristics with other innexin proteins, such as the typical four transmembrane α-helical spanners, two extracellular loops, one intracellular loop, the N-terminal and the C-terminal in the cytoplasm. The C-terminal was the most variable region, either for the length of C-terminal or the amino acid composition. A pair of cysteines residues in each extracellular domain could form the disulfide bonds bridges inside the innexin protein to form the correct polymer for an essential function, possibly as a receptor for specific protein-protein interactions (Kumar and Gilula, 1996; Yeager et al., 1998; Hua et al., 2003). Moreover, the proline residue in the second transmembrane domain is conserved as it was an important component of a molecular hinge motif which acts as a conformational switch responsed to voltage variation (Sansom and Weinstein, 2000; Wang et al., 2015).
It has been reported that arthropod innexins originated from a gene duplication of an ancestral ecdysozoan innexin. This ancestral innexin underwent gene duplication and sequence divergence, which led to different innexins before the separation of insects and crustaceans within the arthropods (Ducret et al., 2006). Phylogeny analysis suggests that the occurrence of Sp-inx1, Sp-inx2, and Sp-inx3 gene may result from a common origin by an intragenic duplication event. Furthermore, it reveals that innexin1 gene occurred earlier than innexin2 and innexin3 genes in arthropods. This is different from the evolution of vertebrate pannexins during which gene duplication occurred leading to the first appearance of pannexin2, and afterwards pannexin1 and pannexin3 that originated from a common ancestor (Abascal and Zardoya, 2013).
Transcription analysis showed that Sp-inx3 mRNA was expressed in all the examined tissues except for the hemocytes and was highly expressed in nervous system tissues including the eyestalk, brain, and thoracic ganglion mass. Moreover, the Sp-inx3 protein was also predominantly expressed in the eyestalk, brain, and thoracic ganglion mass. IHC analysis of Sp-inx3 in the brain and thoracic ganglions mass of adult crabs also confirmed the wide distribution of the Sp-inx3 protein in these two tissues. The eyestalk has a visual function and plays a role in regulating the nervous system and hormones (Bao et al., 2015). Curtin et al. (2002) revealed that the development of neural function in the Drosophila visual system required innexin proteins. Thoracic ganglion mass is one of the nervous organs in S. paramamosain and is involved in the regulation of crab neuroendocrine (Ye et al., 2008). Ducret et al. (2006) have reported that lobster neurons lacking innexin expression displayed weak electrical coupling or dye coupling. From the above, the substantial existence of Sp-inx3 mRNA and protein in the nervous system of S. paramamosain indicates that Sp-inx3 protein may play a role in the nervous system.
To explore the possible role of Sp-inx3 gene in embryonic development, the transcription of Sp-inx3 gene at various development stages were tested. Results show that Sp-inx3 mRNA expressed at a high level from the embryo1 to embryo4 stages, but its transcription decreased at the pre-hatching stages and zoea 1 larvae stage. Previous studies have shown that gap junction coupling is generally widely distributed and highly regulated in the early embryo stages with at least three innexins expressed at the 2-cell stage of C. elegans (Starich et al., 2001, 2003). Ducret et al. (2006) found that innexins of lobster are expressed within the developing nervous system and undergo a marked down-regulation within the stomatogastric ganglion throughout development. There is also a report showing that innexin3 gene of C. elegans is highly expressed in embryo development and exhibits low expression levels in the post-embryonic stage (Starich et al., 2003). However, Sp-inx2 mRNA transcription gradually increases along with the development process. Therefore, the specific function of Sp-inx3 gene is different from Sp-inx2 in the process of embryonic development.
5 CONCLUSIONIn conclusion, our findings suggest that (1) gene duplication events might give rise to Sp-inx1, Sp-inx2, and Sp-inx3; (2) a high level of Sp-inx3 mRNA and protein in eyestalks, brain, and thoracic ganglion mass of adult crabs suggests its important role in the nervous system; and (3) the expression of Sp-inx3 mRNA occurs in two phases with high expression in the embryonic stage and a marked down-regulation after the pre-hatching period. In the future, some other physiological approaches, such as in-situ hybridization and RNAi, is necessary to be used to further study the functional role of Sp-inx3 in embryonic development and the nervous system.
6 DATA AVAILABILITY STATEMENTThe data generated or analyzed during the current study are available from the corresponding author on reasonable request.
Abascal F, Zardoya R. 2013. Evolutionary analyses of gap junction protein families. Biochimica et Biophysica Acta(BBA)-Biomembranes, 1828(1): 4-14.
DOI:10.1016/j.bbamem.2012.02.007 |
Altun Z F, Chen B J, Wang Z W, Hall D H. 2009. High resolution map of Caenorhabditis elegans gap junction proteins. Developmental Dynamics, 238(8): 1936-1950.
DOI:10.1002/dvdy.22025 |
Bao C C, Yang Y A, Huang H Y, Ye H H. 2015. Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain:transcriptomic analysis and expression profiles during vitellogenesis. Scientific Reports, 5: 17055.
DOI:10.1038/srep17055 |
Bauer R, Lehmann C, Fuss B, Eckardt F, Hoch M. 2002. The Drosophila gap junction channel gene innexin 2 controls foregut development in response to Wingless signalling. Journal of Cell Science, 115(Pt 9): 1859-1867.
|
Bauer R, Lehmann C, Martini J, Eckardt F, Hoch M. 2004. Gap junction channel protein innexin 2 is essential for epithelial morphogenesis in the Drosophila embryo. Molecular Biology of the Cell, 15(6): 2992-3004.
DOI:10.1091/mbc.e04-01-0056 |
Bohrmann J, Zimmermann J. 2008. Gap junctions in the ovary of Drosophila melanogaster:localization of innexins 1, 2, 3 and 4 and evidence for intercellular communication via innexin-2 containing channels. BMC Developmental Biology, 8(1): 111.
DOI:10.1186/1471-213X-8-111 |
Bosco D, Haefliger J A, Meda P. 2011. Connexins:key mediators of endocrine function. Physiological Reviews, 91(4): 1393-1445.
DOI:10.1152/physrev.00027.2010 |
Calkins T L, Woods-Acevedo M A, Hildebrandt O, Piermarini P M. 2015. The molecular and immunochemical expression of innexins in the yellow fever mosquito, Aedes aegypti:Insights into putative life stage-and tissuespecific functions of gap junctions. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 183: 11-21.
DOI:10.1016/j.cbpb.2014.11.013 |
Chen F Y, Bo J, Ma X W, Dong L X, Shan Z G, Cui Q, Chen H Y, Wang K J. 2015. A new membrane lipid raft gene SpFLT-1 facilitating the endocytosis of Vibrio alginolyticus in the crab Scylla paramamosain. PLoS One, 10(7): e0133443.
DOI:10.1371/journal.pone.0133443 |
Chuang C F, VanHoven M K, Fetter R D, Verselis V K, Bargmann C I. 2007. An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell, 129(4): 787-799.
DOI:10.1016/j.cell.2007.02.052 |
Curtin K D, Zhang Z, Wyman R J. 2002. Gap junction proteins expressed during development are required for adult neural function in the Drosophila optic lamina. Journal of Neuroscience, 22(16): 7088-7096.
DOI:10.1523/JNEUROSCI.22-16-07088.2002 |
Ducret E, Alexopoulos H, Le Feuvre Y, Davies J A, Meyrand P, Bacon J P, Fénelon V S. 2006. Innexins in the lobster stomatogastric nervous system:cloning, phylogenetic analysis, developmental changes and expression within adult identified dye and electrically coupled neurons. European Journal of Neuroscience, 24(11): 3119-3133.
DOI:10.1111/j.1460-9568.2006.05209.x |
Dykes I M, Macagno E R. 2006. Molecular characterization and embryonic expression of innexins in the leech Hirudo medicinalis. Development Genes and Evolution, 216(4): 185-197.
DOI:10.1007/s00427-005-0048-1 |
Ganfornina M D, Sánchez D, Herrera M, Bastiani M J. 1999. Developmental expression and molecular characterization of two gap junction channel proteins expressed during embryogenesis in the grasshopper Schistocerca americana. The Journal of Genetics and Development, 24(1-2): 137-150.
DOI:10.1002/(SICI)1520-6408(1999)24:1/2<137::AID-DVG13>3.0.CO;2-7 |
Giuliani F, Giuliani G, Bauer R, Rabouille C. 2013. Innexin 3, a new gene required for dorsal closure in Drosophila embryo. PLoS One, 8(7): e69212.
DOI:10.1371/journal.pone.0069212 |
Holcroft C E, Jackson W D, Lin W H, Bassiri K, Baines R A, Phelan P. 2013. Innexins Ogre and Inx2 are required in glial cells for normal postembryonic development of the Drosophila central nervous system. Journal of Cell Science, 126(17): 3823-3834.
DOI:10.1242/jcs.117994 |
Hong S M, Kang S W, Goo T W, Kim N S, Leed J S, Kima K A, Nho S K. 2008. Two gap junction channel (innexin)genes of the Bombyx mori and their expression. Journal of Insect Physiology, 54(1): 180-191.
DOI:10.1016/j.jinsphys.2007.09.002 |
Hua V B, Chang A B, Tchieu J H, Kumar N M, Nielsen P A, Saier Jr M H. 2003. Sequence and phylogenetic analyses of 4 TMS junctional proteins of animals:connexins, innexins, claudins and occludins. The Journal of Membrane Biology, 194(1): 59-76.
DOI:10.1007/s00232-003-2026-8 |
Kandarian B, Sethi J, Wu A, Baker M, Yazdani N, Kym E, Sanchez A, Edsall L, Gaasterland T, Macagno E. 2012. The medicinal leech genome encodes 21 innexin genes:different combinations are expressed by identified central neurons. Development Genes and Evolution, 222(1): 29-44.
DOI:10.1007/s00427-011-0387-z |
Kielian T. 2008. Glial connexins and gap junctions in CNS inflammation and disease. Journal of Neurochemistry, 106(3): 1000-1016.
DOI:10.1111/j.1471-4159.2008.05405.x |
Kumar N M, Gilula N B. 1996. The gap junction communication channel. Cell, 84(3): 381-388.
DOI:10.1016/S0092-8674(00)81282-9 |
Lechner H, Josten F, Fuss B, Bauer R, Hoch M. 2007. Cross regulation of intercellular gap junction communication and paracrine signaling pathways during organogenesis in Drosophila. Developmental Biology, 310(1): 23-34.
DOI:10.1016/j.ydbio.2007.07.008 |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4): 402-408.
DOI:10.1006/meth.2001.1262 |
Lopresti V, Macagno E R, Levinthal C. 1974. Structure and development of neuronal connections in isogenic organisms:transient gap junctions between growing optic axons and lamina neuroblasts. Proceedings of the National Academy of Sciences of the United States of America, 71(4): 1098-1102.
DOI:10.1073/pnas.71.4.1098 |
McCracken C B, Roberts D C S. 2006. Neuronal gap junctions:expression, function, and implications for behavior. International Review of Neurobiology, 73: 125-151.
DOI:10.1016/S0074-7742(06)73004-5 |
Nagy J I, Dudek F E, Rash J E. 2004. Update on connexins and gap junctions in neurons and glia in the mammalian nervous system. Brain Research Reviews, 47(1-3): 191-215.
DOI:10.1016/j.brainresrev.2004.05.005 |
Northcutt A J, Lett K M, Garcia V B, Diester C M, Lane B J, Marder E, Schulz D J. 2016. Deep sequencing of transcriptomes from the nervous systems of two decapod crustaceans to characterize genes important for neural circuit function and modulation. BMC Genomics, 17(1): 868.
DOI:10.1186/s12864-016-3215-z |
Oshima A, Matsuzawa T, Nishikawa K, Fujiyoshi Y. 2013. Oligomeric structure and functional characterization of Caenorhabditis elegans innexin-6 gap junction protein. Journal of Biological Chemistry, 288(15): 10513-10521.
DOI:10.1074/jbc.M112.428383 |
Oshima A, Tani K, Fujiyoshi Y. 2016. Atomic structure of the innexin-6 gap junction channel determined by cryo-EM. Nature Communications, 7: 13681.
DOI:10.1038/ncomms13681 |
Phelan P, Goulding L A, Tam J L Y, Allen M J, Dawber R J, Davies J A, Bacon J P. 2008. Molecular mechanism of rectification at identified electrical synapses in the Drosophila giant fiber system. Current Biology, 18(24): 1955-1960.
DOI:10.1016/j.cub.2008.10.067 |
Phelan P, Stebbings L A, Baines R A, Bacon J P, Davies J A, Ford C. 1998. Drosophila Shaking-B protein forms gap junctions in paired Xenopus oocytes. Nature, 391(6663): 181-184.
DOI:10.1038/34426 |
Phelan P. 2005. Innexins:members of an evolutionarily conserved family of gap-junction proteins. Biochimica et Biophysica Acta (BBA)-Biomembranes, 1711(2): 225-245.
DOI:10.1016/j.bbamem.2004.10.004 |
Sansom M S P, Weinstein H. 2000. Hinges, swivels and switches:the role of prolines in signalling via transmembrane α-helices. Trends in Pharmacological Sciences, 21(11): 445-451.
DOI:10.1016/S0165-6147(00)01553-4 |
Shruti S, Schulz D J, Lett K M, Marder E. 2014. Electrical coupling and innexin expression in the stomatogastric ganglion of the crab Cancer borealis. Journal of Neurophysiology, 112(11): 2946-2958.
DOI:10.1152/jn.00536.2014 |
Skerrett I M, Williams J B. 2017. A structural and functional comparison of gap junction channels composed of connexins and innexins. Developmental Neurobiology, 77(5): 522-547.
DOI:10.1002/dneu.22447 |
Starich T A, Miller A, Nguyen R L, Hall D H, Shaw J E. 2003. The Caenorhabditis elegans innexin INX-3 is localized to gap junctions and is essential for embryonic development. Developmental Biology, 256(2): 403-417.
DOI:10.1016/S0012-1606(02)00116-1 |
Starich T, Sheehan M, Jadrich J, Shaw J. 2001. Innexins in C. elegans. Cell Communication & Adhesion, 8(4-6): 311-314.
DOI:10.3109/15419060109080744 |
Stebbings L A, Todman M G, Phillips R, Greer C E, Tam J, Phelan P, Jacobs K, Bacon J P, Davies J A. 2002. Gap junctions in Drosophila:developmental expression of the entire innexin gene family. Mechanisms of Development, 113(2): 197-205.
DOI:10.1016/S0925-4773(02)00025-4 |
Wang S P, Chen F Y, Dong L X, Zhang Y Q, Chen H Y, Qiao K, Wang K J. 2015. A novel innexin2 forming membrane hemichannel exhibits immune responses and cell apoptosis in Scylla paramamosain. Fish & Shellfish Immunology, 47(1): 485-499.
DOI:10.1016/j.fsi.2015.09.028 |
Watanabe T, Kankel D R. 1992. The l(1)ogre gene of Drosophila melanogaster is expressed in postembryonic neuroblasts. Developmental Biology, 152(1): 172-183.
DOI:10.1016/0012-1606(92)90167-F |
Wu C L, Shih M F M, Lai J S Y, Yang H T, Turner G C, Chen L Y, Chiang A S. 2011. Heterotypic gap junctions between two neurons in the Drosophila brain are critical for memory. Current Biology, 21(10): 848-854.
DOI:10.1016/j.cub.2011.02.041 |
Ye H H, Huang H Y, Li S J, Wang G Z. 2008. Immunorecognition of estrogen and androgen receptors in the brain and thoracic ganglion mass of mud crab, Scylla paramamosain. Progress in Natural Science, 18(6): 691-695.
DOI:10.1016/j.pnsc.2007.12.012 |
Yeager M, Unger V M, Falk M M. 1998. Synthesis, assembly and structure of gap junction intercellular channels. Current Opinion in Structural Biology, 8(4): 517-524.
DOI:10.1016/S0959-440X(98)80131-0 |
Yen M R, Saier Jr M H. 2007. Gap junctional proteins of animals:the innexin/pannexin superfamily. Progress in Biophysics and Molecular Biology, 94(1-2): 5-14.
DOI:10.1016/j.pbiomolbio.2007.03.006 |
 2019, Vol. 37
2019, Vol. 37


