Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LI Zan, LIU Xiumei, DU Xinxin, ZHANG Kai, CHEN Yan, WANG Xubo, WANG Zhigang, YU Haiyang, ZHANG Quanqi
- Tumor necrosis factor-alpha (TNF-α) in spotted halibut Verasper variegatus at the embryonic and metamorphic stages
- Journal of Oceanology and Limnology, 38(2): 454-466
- http://dx.doi.org/10.1007/s00343-019-8289-4
Article History
- Received Nov. 8, 2018
- accepted in principle Jan. 28, 2019
- accepted for publication Jun. 30, 2019
2 College of Life Sciences, Yantai University, Yantai 264005, China;
3 Key Laboratory of Marine Genetics and Breeding, Ministry of Education, College of Marine Life Science, Ocean University of China, Qingdao 266003, China
Tumor necrosis factor-alpha (TNF-α) belongs to the "TNF superfamily", a large family of structurally related proteins. TNF, a pleiotropic cytokine, is critical to the control of an extensive array of immunological responses of monocytes, macrophages, T- and B-lymphocytes, and NK cells (Sherry and Cerami, 1988; Camussi et al., 1991; Nascimento et al., 2007; Li and Zhang, 2016). It can increase cell survival, induce cellular differentiation, apoptosis, and necrosis, and contribute to both physiological and pathological processes (Li and Zhang, 2016). Furthermore, TNF-α can serve as a signaling molecule and interact with its receptors TNFR-1 and TNFR-2 to transduce the exterior signals into the cells (Loetscher et al., 1991; Idriss and Naismith, 2000; Li and Zhang, 2016; Qi et al., 2016).
The functions of TNF-α are manifold (Beg and Baltimore, 1996; Blobel, 1997; Uysal et al., 1997; Idriss and Naismith, 2000; Ellis, 2001; Locksley et al., 2001). First, this protein can confer resistance to infections. TNF-α receptor knockout leads to less resistance against pathogens and weaken the inflammatory response (Acton et al., 1996). Second, TNF-α has also been reported to be related to the embryonic development process (Wride and Sanders, 1995). Third, TNF-α is involved in physiological sleep regulation in animal models (Idriss and Naismith, 2000). Fourth, TNF-α can regulate apoptosis either during normal embryonic development or during organelle destruction (Wride and Sanders, 1995; Idriss and Naismith, 2000). These studies show that TNF-α is a crucial signaling protein during an immune reaction, and it is also involved in developmental and other biological processes.
TNF-α was first cloned in humans by Pennica et al. (1984). It has been identified and characterized in teleosts, such as Cynoglossus semilaevis (Li and Zhang, 2016), Paralichthys olivaceus (Hirono et al., 2000), Oncorhynchus mykiss (Zou et al., 2002; Hong et al., 2013), Scophthalmus maximus (Ordás et al., 2007), Sparus aurata (García-Castillo et al., 2002; Saeij et al., 2003), Ctenopharyngodon idella (Zhang et al., 2012), Cyprinus carpio (Forlenza et al., 2009), Oplegnathus fasciatus (Hwang et al., 2014), Dicentrarchus labrax (Nascimento et al., 2007), Siniperca chuatsi (Xiao et al., 2007), Carassius auratus (Grayfer et al., 2008), and Ictalurus punctatus (Zou et al., 2003). TNF-α in C. idella was found involved in the regulation of the NF-κB pathway (Zhang et al., 2012). In O. mykiss, two types of TNF-α were found, of which TNF-α3 could induce the expression of macrophage growth factor, antimicrobial peptides, and proinflammatory cytokines (Hong et al., 2013). The expression of I. punctatus TNF-α was found up-regulated in the peripheral blood leukocytes (PBLs) by phorbol-12-myristate-13-acetate (PMA)/ calcium ionophore treatment (Zou et al., 2003). Given that TNF-α is generally regarded as an immunity gene, many researchers have studied the immunological function of TNF-α in fishes. However, the role of TNF-α in cellular differentiation and apoptosis in fishes has been rarely studied.
As V. variegatus has a high market value in Asia, it is widely considered a promising candidate for aquaculture and fishery enhancement. However, there are several problems in the current culture process of V. variegatus. In the process of artificial breeding, we have not found an effective way to make its broodfish mature and ovulate spontaneously (Xu et al., 2012). In addition, V. variegatus is susceptible to virus and bacterial infections, which makes it endangered. Thus, the function studies of immunity genes could help us to find effective methods to improve the survival rate in its aquaculture. TNF-α has not been identified, and its function remains unclear in V. variegatus. To better understand the function of TNF-α gene in V. variegatus (VvTNF-α), we first cloned its full-length cDNA and predicted the secondary and 3D structure of VvTNF-α protein. Then we studied its expression pattern in different adult tissues and during embryonic development, including metamorphosis. Additionally, we chose the PBLs of V. variegatus to detect the immune challenge response stimulated by pathogen-associated molecular patterns (PAMPs). The above data are essential for us to understand the functions of VvTNF-α in immune response and its novel functions during the metamorphic stages.
2 MATERIAL AND METHOD 2.1 Ethics statementVerasper variegatus samples were obtained from a commercial hatchery. This research was conducted in accordance with the protocols of the Institutional Animal Care and Use Committee of the Ocean University of China (protocol number 11-06) and the China Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (State Science and Technology Commission of the People's Republic of China for No. 2, October 31, 1988. http://www.gov.cn/gongbao/content/2011/content_1860757.htm).
2.2 Sample collectionThe V. variegatus used in this study were raised in a commercial hatchery in Weihai, Shandong Province, China. The fish were anesthetized (MS-222 at 30 μg/mL) and then killed by severing spinal cord. To research the expression pattern in different tissues, heart, liver, spleen, kidney, brain, gill, muscle, intestine and PBLs were collected from three 1-yearold samples. Each of these samples was collected in triplicate. Samples were snap frozen in liquid nitrogen and then stored at -80℃ until further use. Fertilized eggs were released by artificial fertilization. The eggs and larvae were incubated and reared in plastic tanks (length: 94.6 cm; width: 65.1 cm; height: 49.7 cm; volume: 0.306 m3; fish density: 2.3 kg/m3) with aerated seawater (dissolved oxygen: 4.6 mg/L; salinity: 30; pH value: 7.9; light intensity: 800 lx; temperature: 11±1℃) for several days before random sampling. The different embryonic stages were observed under a stereomicroscope. Three pools of samples at 10 embryonic stages and 6 metamorphic stages were separately collected from mixed families with a nylon net (100 mesh). The embryonic stages includes unfertilized egg, 1-cell, 4-cell, morula, blastula, gastrula, neurula, somite, tail bud, and hatching stages. And the metamorphic stages includes 3 days post hatching, 10 days post hatching, premetamorphosis, early-metamorphosis, midmetamorphosis, late-metamorphosis. The above samples were immersed in 1.5 mL of RNAwait liquid (Solarbio, Shanghai, China) overnight at 4℃ and then stored at -80℃ until further use.
2.3 RNA extraction and cDNA synthesisAs per the manufacturer's instructions, we used TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) to extract total RNA. The quality and quantity of the total RNA were evaluated by 1.5% agarose gel electrophoresis and spectrophotometry with the NanoPhotometer Pearl (Thermo Scientific, Carlsbad, CA, USA). We used RNase-free DNase Ⅰ (TaKaRa, Dalian, China) to remove the DNA contamination of the extracted total RNA and then frozen at -80℃.
As per the manufacturer's protocol of M-MLV kit (TaKaRa, Dalian, China), reverse transcription and cDNA synthesis were performed with 1 μg of the above RNA and using random hexamer primers.
2.4 Molecular cloning and sequence analysis of VvTNF-αThe genome information of V. variegatus has not yet been available. To obtain the conserved region of VvTNF-α, we designed a pair of degenerate primers (TNF-Fw/Rv, Table 1) in accordance with the conserved sequences of TNF-α in other teleosts. The species and GenBank accession numbers utilized as reference to the construction of the degenerated primers were as follows: P. olivaceus TNF-α (BAA94969.1), P. maxima TNF-α (ACN41911.1), P. major TNF-α (AAP76392.1) and D. labrax TNF-α (AAZ20770.1). The PCR reaction system in the above experiment used to obtain the open reading frame (ORF) sequence was as follows: 2.5 μL 10×buffer (15 mmol/L), 0.5 μL dNTP (10 mmol/L), 0.5 μL Primer-Fw (10 mmol/L), 0.5 μL Primer-Rv (10 mmol/L), 1 μL cDNA template (20 ng/μL), 0.25 μL Taq DNA polymerase (5 U/μL), and 19.75 μL sterile water. PCR amplification program was as follows: 95℃ for 5 min; 30 cycles of 95℃ for 30 s, 55℃ for 30 s, and 72℃ for 1 min; and 72℃ for 15 min. To obtain the TNF-α full-length cDNA from the spleen sample of V. variegatus, we performed the 5′- and 3′-rapid amplification of cDNA ends (RACE) by using the SMART RACE cDNA Amplification Kit (Clontech, CA, USA) based on the manufacturer's instruction. The gene-specific primers (GSPs) for the nested PCR assay were TNF-5′Rv1/Rv2 for the 5′- RACE and TNF-3′Fw1/Fw2 for the 3′-RACE (Table 1). The PCR was conducted in accordance with the SMART RACE amplification protocol. We separated the PCR products by using 1.5% agarose gel electrophoresis. The separated PCR products were purified by using the Zymoclean Gel DNA Recovery Kit (Zymo Research, CA, USA) and cloned into the pMD-18T vector (TaKaRa) according to the manufacturer's instruction, and then the above products were sequenced.
ORF of VvTNF-α was predicted by using the ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
2.5 Quantitative real-time PCR (qRT-PCR)TNF-α-RT-FW/RV (Table 1, amplification efficiency=0.99), the specific primer pair used in this experiment, was designed according to the characterized of TNF-α. Pre-experiments with qRTPCR amplification using specific primers and spleen cDNA template were conducted to confirm whether the melt curve was unimodal. The unimodal melt curve corresponded to a single cDNA qRT-PCR products that could be used for subsequent experiments. As 18S rRNA was the most stable reference gene in different tissues and at different developmental stages, it was used as the reference gene in the relative expression analysis of VvTNF-α.
Three biological replicates of each sample were analyzed, with each sample ran in triplicate. The qRTPCR reaction system was as follows: 1 μL cDNA template (10 ng/μL), 0.4 μL Primer-Fw (10 mmol/L), 0.4 μL Primer-Rv (10 mmol/L), 8.2 μL sterile water, and 10 μL SYBR Premix Ex Taq Ⅱ (TaKaRa). qRTPCR was performed by using LightCycler 480 at 95℃ for 5 min pre-incubation, followed by 45 cycles of 95℃ for 15 s and 60℃ for 45 s. We detected single amplification by analyzing the melting curve. Under the control of LightCycler 480 Software 1.5, fluorescent signal accumulation was recorded at the 60℃ 45 s phase during each cycle. We used the 2-ΔΔCt comparative Ct method to calculate the relative quantities of VvTNF-α expressed as fold variation over reference gene.
2.6 PAMPs-induced VvTNF-α expression in PBLsWe collected the blood from the caudal veins of the V. variegatus samples, and preared the PBLs using Percoll as previously reported (Zhou et al., 2014). The trypan blue exclusion method was used to determine the viability of PBLs and the average percent viability of PBLs is 93.6%. After full mixing, V. variegatus PBLs were cultured in a 24-well plate (Thermal Scientific, 9×106 cells/well) overnight at 24℃. And then the V. variegatus PBLs were separately stimulated with lipopolysaccharide (LPS) (50 μg/mL), polyinosinic: polycytidylic acid (poly(I:C), Sigma) (50 μg/mL), and PBS (control). After 0, 0.5, 1, 2, 4, 6, 12, and 24 h of PAMPs treatment, the samples were separately obtained. In order to study the response of VvTNF-α to LPS, poly(I:C), and PBS challenge, the qRT-PCR was constructed using 18S rRNA as reference gene. The above experiment was repeated three times, so three biological replicates of each sample were analyzed, with each sample ran in triplicate.
2.7 Bioinformatics analysis and phylogenetic tree reconstructionThe homologous nucleotide and protein sequences of VvTNF-α were confirmed by a BLAST search against the databases of NCBI and Ensembl. Multiple sequence alignments were performed with ClustalX 2.1 and DNAMAN 7.0. The phylogenetic tree was constructed through MrBayes 3.2.3.
The motifs of vertebrate TNF-α sequences were searched using the Pfam and CCD databases. Use Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) and PDBsum Generate (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html) to predict the secondary and 3D structure of protein VvTNF-α. The docking simulation of protein was performed with the ZDOCK server (http://zdock.umassmed.edu/).
2.8 Statistical analysisUse a one-way ANOVA to analyze qRT-PCR data statistically followed by LSD test by SPSS 20.0 (IBM, New York, USA). P < 0.05 is considered statistically significant. The data are showed as mean±SD (n=3).
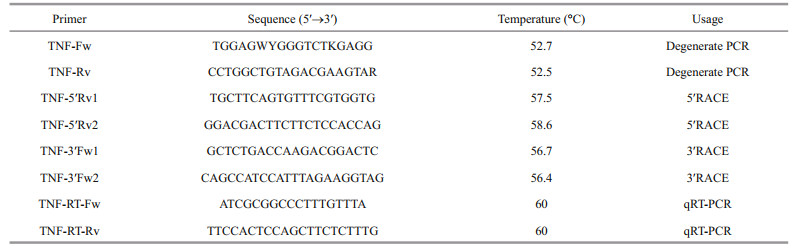
3 RESULT AND DISCUSSION 3.1 VvTNF-α is highly conserved among vertebratesThe V. variegatus genome information has not yet been available. Thus, we designed a pair of degenerate primers (TNF-Fw/Rv, Table 1) in accordance with the conserved sequences of other vertebrates TNF-α and gained the conserved region of VvTNF-α. Then, we used the RACE method to obtain the unknown 5′- and 3′-regions of VvTNF-α cDNA. To obtain the VvTNF-α cDNA sequence, we assembled all the cloned sequences using SeqMan. The full-length cDNA of VvTNF-α is 1 273-bp long (GenBank: KY038170) and includes a 100-bp 5′-untranslated region (UTR), a 417-bp 3′-UTR with a poly(A) tail, and a 756-bp ORF. VvTNF-α has a complete polyadenylation signal in the 3′-UTR in comparison with P. olivaceus TNF-α. In addition, TA-rich motifs (TTATTTAT) in the 3′-UTR of mammalian TNF-α are also present in the VvTNF-α cDNA. These motifs influence the half-life (Caput et al., 1986) and translational efficiency (Han et al., 1990) of TNF-α mRNA. The 3′-UTRs of human and mouse TNF, as well as those of human lymphotoxin, human CSF, human and rat fibronectin, human and mouse IL-1, and a majority of sequenced human and mouse IFNs, have the consensus sequence TTATTTAT (Caput et al., 1986). Except for lymphotoxin mRNA, all of these mRNAs have no homology to the coding region of TNF mRNAs (Caput et al., 1986). One of the characteristic feature of inflammatory mediator genes is the TTATTTAT instability motif (Sachs, 1993), whose presence shows the VvTNF-α transient expression. TA-rich motifs are common in sequences encoding inflammatory response-related proteins (Caput et al., 1986).
The ORF of VvTNF-α encoded a polypeptide comprising 251 amino acid residues, whose molecular weight is 27.94 kDa and theoretical isoelectric point is 5.57. Like other vertebrates, there was a transmembrane (TM) domain (residues numbers 37– 54) at the N-terminus of VvTNF-α predicted by Singer's classification for membrane topology (Singer, 1990). The predicted VvTNF-α polypeptide also contains the TNF domain in the C-terminus of the protein. The sequence identities of VvTNF-α with the TNF-α of S. maximus, O. fasciatus, P. olivaceus, and Homo sapiens are 83%, 82%, 81%, and 54%, respectively. These results imply that the TNF-α protein may be highly conserved among vertebrates.
3.2 Phylogenetic and protein domains of VvTNF-α are consistent with those of other teleosts TNF-αThe alignment of VvTNF-α with the other vertebrates TNF-α shows that the VvTNF-α contains all the basic elements of the TNF gene family characteristic in the fish and higher vertebrate. These elements include the TNF family signature [VL]-x- [LIVM]-x3-G-[LIVMF]-Y-[LIVMFY]2-x2-[QEKHL] (with the exception of I instead of V or L at position 1), a TM domain, and two conserved cysteine residues, which are critical for the correct folding of mature TNF-α (Rink and Kirchner, 1996) (Fig. 1a). As shown in Fig. 1a, like other fish TNF-α, the VvTNF-α also contains the Thr-Leu motif which could be recognized by the TACE metalloproteinase. In soluble mouse TNF-α, the Thr-Leu motif was confirmed as the cleavage site for the release of mature protein (McGeehan et al., 1994). Moreover, most sequence identities are located at the C-terminus domain, which is consistent with the other teleosts TNF protein (Praveen et al., 2006) (Fig. 1a).
|
| Fig.1 The amino acid sequence alignment and phylogenetic tree analysis of vertebrates TNF a. the alignment of fishes and mammals TNFs through ClustalW method. Asterisks, colons, and dots indicate different degree of conserved residues, respectively. The TNF family signature is boxed, the putative TACE cleavage sites are in red letters and the two conserved cysteines are directed by two red arrows; b. phylogenetic analysis showing the relationship between VvTNF-α and other vertebrates TNF-α. A phylogram was constructed through MyBayes (mcmc=200 000 generations, sample freq.=10). H. sapiens TNF-β served as the out-group. The species and GenBank accession numbers were as follows: H. sapiens TNF-α (NP_000585.2); M. musculus TNF-α (AAI37721.1); A. sinensis TNF-α (XP_006036188.1); X. tropicalis TNF-α (NP_001107143.1); P. olivaceus TNF-α (BAA94969.1); P. maxima TNF-α (ACN41911.1); D. labrax TNF-α (AAZ20770.1); E. coioides TNF-α (ACM45963.1); P. major TNF-α (AAP76392.1); T. rubripes TNF-α (NP_001033074.1); O. niloticus TNF-α (NP_001266462.1); M. zebra TNF-α (XP_004573618.1); O. mykiss TNF-α (CAC16408.1); H. sapiens TNF-β (AAB59455.1). The TNF and other domains of TNFs were showed with the CDD server (Marchler-Bauer and Bryant, 2004; Marchler-Bauer et al., 2009, 2011, 2015) |
A search for all the proteins against the Pfam database (http://pfam.xfam.org/) revealed that each of these proteins contains one TNF-α domain located at the C-terminus of the protein (Fig. 1b). As shown in Fig. 1b, the vertebrate TNF-α protein phylogenetic tree constructed with MrBayes shows that TNF-α is clustered into one group but separated from the outgroup H. sapiens TNF-β. This indicates that the production of the TNF-α and TNF-β is due to TNF ancestor gene duplication, which occurred before the species formation. The fishes TNF-α are clustered, and the TNF-α of V. variegatus, P. maxima, and P. olivaceus are clustered into one group.
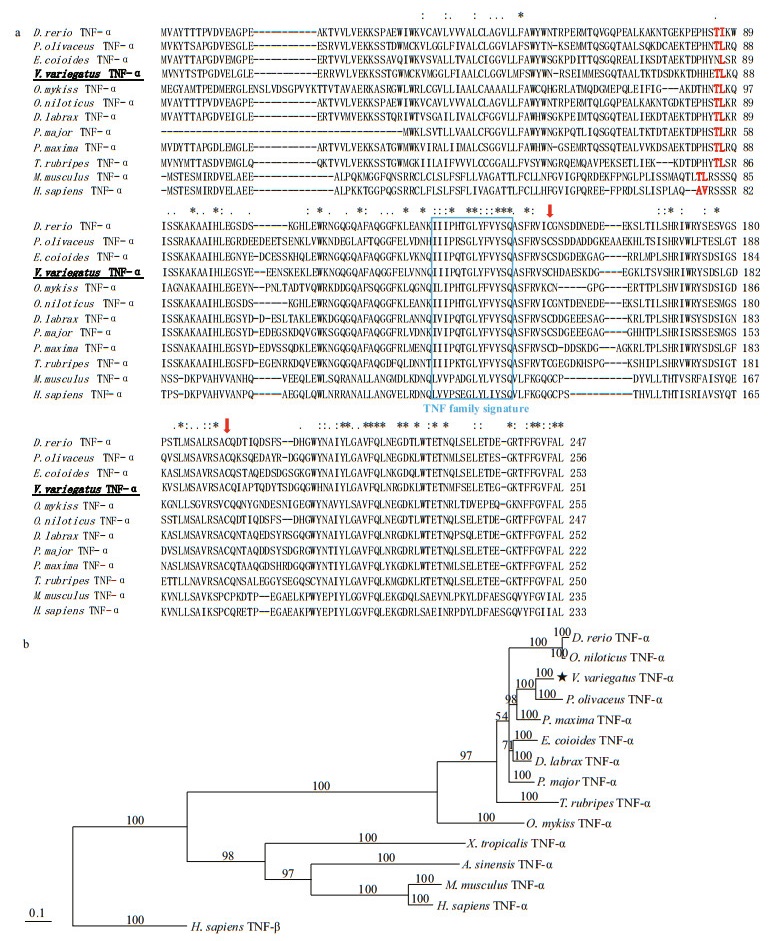
3.3 VvTNF-α is highly expressed in the PBLs, gill, spleen and at the hatching stageThe expression of TNF-α can be regulated (Hawiger, 2001); thus, we explored the expression patterns of VvTNF-α in different tissues and at different embryonic developmental stages with qRTPCR. The VvTNF-α expression level was normalized to the housekeeping genes, 18S rRNA. As shown in Fig. 2, VvTNF-α is widely expressed, which is consistent with the tissue distribution results for C. carpio, I. punctatus, S. aurata, and D. labrax (Zou et al., 2003; García-Castillo et al., 2004; Savan and Sakai, 2004; Nascimento et al., 2007). The presence of the VvTNF-α transcript in the brain is of interest, given that TNF-α is believed to be involved in the physiological sleep regulation in mammals (García-Castillo et al., 2002). However, our result shows that, VvTNF-α had the least amount of expression in the brain. The VvTNF-α transcripts are most expressed in the PBLs, followed by the spleen and gill. The other tissues, such as liver, heart, kidney, muscle, and intestine show moderate level of VvTNF-α transcripts. This result is similar to the phenomenon in C. auratus, S. aurata, I. punctatus, and O. mykiss (García-Castillo et al., 2002; Zou et al., 2003; Grayfer et al., 2008; Hong et al., 2013).
|
| Fig.2 Relative expression of VvTNF-α in different adult tissues The relative expression variance is represented by a ratio (amount of VvTNF-α normalized to the values of 18S rRNA). The relative expression level in brain is normalized to 1. The data are shown as mean±SD (n=3). Columns with different letters show a significant difference (P < 0.05). |
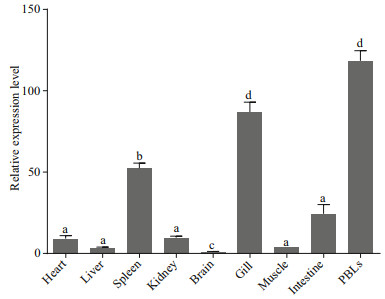
In the early stages of embryonic development, the egg envelope protects the fish embryo from the attacks by pathogens in the water environment. After hatching, the fish embryos exposed to the environment are vulnerable to pathogen. At present, we know very little about the immune defense mechanism of fish embryo in their hostile environment. Thus, we explored the temporal expression pattern of VvTNF-α during the embryonic stages. The result showed that VvTNF-α mRNA was detected until the somitestage (Fig. 3). This phenomenon indicates that VvTNF-α is not a maternally expressed gene. During embryonic development, TNF-α may have major roles in the programmed cell death, in the remodeling of the extracellular matrix and in the cellular growth and differentiation (Wride and Sanders, 1995). Furthermore, the embryonic cells differentiate begin at the somite stage. The TNF-α function during the embryonic development may be supported by our results. Before the hatching stage, the VvTNF-α remained relatively low expression. However, VvTNF-α had a high expression level at the hatching stage (Fig. 3). The exposed aquatic environment of most fish embryos and hatchings is full of a large number of different microorganisms, including potential pathogens; and during this period, the immune systems of those fishes have not yet fully developed (Wang et al., 2016). Furthermore, during the hatching stages, embryos are more susceptible to attack by pathogens due to loss of egg envelope. Therefore, high expression of VvTNF-α in the hatching stage indicates that it may plays a role in resisting the pathogens around the environment. However, TNF-α has not yet been identified in the other fish eggs. Thus, the role of TNF-α in early developmental stage of fish remains largely unclear. In this study, we found that VvTNF-α is not a maternal gene, but its transcripts gradually increases and peaks at the hatching stage. The VvTNF-α expression pattern are similar to those of fish-egg lectin (FEL), which plays a critical role in the immunity of zebra fish and rock bream (Kim et al., 2011; Wang et al., 2016).
|
| Fig.3 Relative expression of VvTNF-α at different embryonic developmental stages The relative expression variance is represented by a ratio (amount of VvTNF-α normalized to the values of 18S rRNA). The relative expression level in neurula stage is normalized to 1. The data are shown as mean±SD (n=3). Columns with different letters show a significant difference (P < 0.05).s |
The presented results show that the expression of inflammatory factor VvTNF-α is tissue and stage specific.
3.4 VvTNF-α shows the highest expression at the mid-metamorphosis stageMetamorphosis of fish in which larva transited to juvenile is a crucial development stage. Pleuronectiformes could change from a symmetrical larva to an asymmetrical juvenile during this stage, which is a dramatic morphological reorganization. Thus, Pleuronectiformes can be used as a representative of teleost to study metamorphosis. Also as flatfish, V. variegatus and P. olivaceus are evolutionary relatives (Li et al., 2011, 2012; Xu et al., 2012), and both undergo the metamorphic stage. Metamorphosis arises from a series of processes including apoptosis, biochemical changes and so on (Power et al., 2001).T3 (Thyroid hormones 3), T4 (Thyroid hormones 4), and TR (thyroid hormone receptor) have been reported to drive flatfish metamorphosis (Inui and Miwa, 1985; De Jesus et al., 1991; Yamano et al., 1994; Campinho et al., 2007), which are related to apoptosis and cellular differentiation (Yamano and Miwa, 1998; Power et al., 2001; Liu and Chan, 2002; Marchand et al., 2004; Klaren et al., 2008).
TNF-α gene plays an important role in cellular differentiation, proliferation, and apoptosis (Wride and Sanders, 1995; Li and Zhang, 2016) and interact with T3, T4, and TR (Aust et al., 1996; Kalashnikova et al., 2009; Kiss-Toth et al., 2013). For instance, thyroid modulation is realized as more intense induction of TNF-α apoptosis in the cells (Kalashnikova et al., 2009) and TNF-α in the thyroid can influence its functions (Aust et al., 1996; Kiss-Toth et al., 2013). In V. variegatus development, VvTNF-α expression began to decrease at 3 days post hatching (dph) and gradually increased again during metamorphic stage followed by another decline at the end of metamorphosis (Fig. 4). In this period, VvTNF-α expression was predominantly detected in mid-metamorphosis, which is the most violent period of metamorphosis (Fig. 4). The results show that VvTNF-α has a consistent expression pattern in comparison to T3, T4, and TR at the metamorphic stage of S. maximus and P. olivaceus (Yamano and Miwa, 1998; Marchand et al., 2004). The relative expression level of TR gene and concentration of T3 and T4 in P. olivaceus was gradually growing during pre-metamorphosis, early-metamorphosis, and midmetamorphosis. They reached a peak in midmetamorphosis and then began to decline (Yamano and Miwa, 1998). In summary, our results strongly imply that VvTNF-α may be involved in a series of regulated processes during flatfish metamorphosis. These results contribute to the further understanding of the molecular mechanism of flatfish metamorphosis.
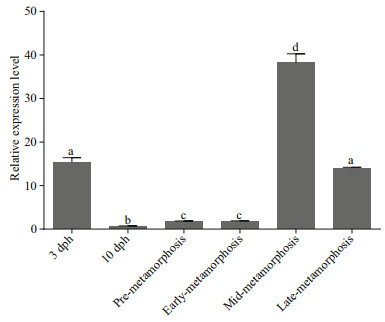
|
| Fig.4 Relative expression of VvTNF-α during metamorphosis The relative expression variance is represented by a ratio (amount of VvTNF-α normalized to the values of 18S rRNA). The relative expression level in 10dph is normalized to 1. The dph stands for days post hatching. The data are shown as mean±SD (n=3). Columns with different letters show a significant difference (P < 0.05). |
As an immunity gene, TNF-α participates in the immune defense against pathogens. Mammalian TNF-α plays a crucial role in the antibacterial immune response induced by Mycobacterium tuberculosis, Listeria monocytogenes and Mycobacterium bovis (Pasparakis et al., 1996; Olleros et al., 2002; Saunders et al., 2005). Because TNF-α is released from macrophages, monocytes, neutrophils, NK cells, and T-cells (Covello et al., 2009), we investigated the VvTNF-α expression patterns in the PBLs exposed to PAMPs. In our study, the V. variegatus PBLs were co-incubated with LPS, poly(I:C) and PBS as a control, respectively. In the qRT-PCR results, we normalized the VvTNF-α expression level to the control. As shown in Figs. 5 and 6, the VvTNF-α fold change levels were significantly and rapidly upregulated at 0.5 h and 1 h after LPS and poly(I:C) treatments, respectively. This result indicates that TNF-α expression may be more sensitive to the PAMPs of LPS than to those of poly(I:C).
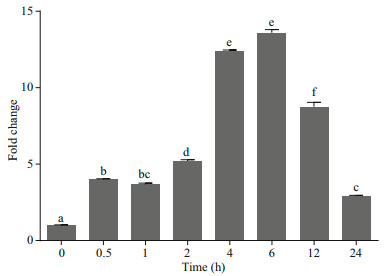
|
| Fig.5 Expression analysis of VvTNF-α in PBLs treated with 50 μg/mL LPS The fold change variance is represented by a ratio (amount of VvTNF-α normalized to the values of 18S rRNA). The time zero is normalized to 1. The data are shown as mean±SD (n=3). Columns with different letters show a significant difference (P < 0.05). |
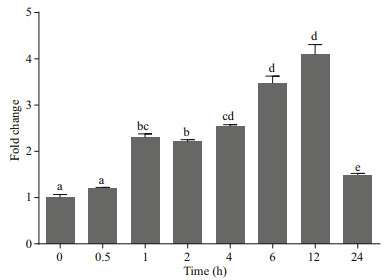
|
| Fig.6 Expression analysis of VvTNF-α in PBLs treated with 50 μg/mL poly(I:C) The fold change variance is represented by a ratio (amount of VvTNF-α normalized to the values of 18S rRNA). The time zero is normalized to 1. The data are shown as mean±SD (n=3). Columns with different letters show a significant difference (P < 0.05) |
TNF-α could be expressed in the human lymphocytesp, monocytes/macrophages and eripheral blood mononuclear cells stimulated by PAMPs (Jang et al., 2006). In addition, reports showed that bacteria, LPS, virus, poly(I:C) and parasites could induce the high expression of TNF-α in a variety of fish (Chang et al., 2006; Roca et al., 2008; Covello et al., 2009; Teles et al., 2011; Hwang et al., 2014). In our study, the VvTNF-α was expressed in various tissues under normal physiological conditions; the VvTNF-α transcripts significantly and rapidly increased after LPS and poly(I:C) challenges. These results indicate VvTNF-α is involved in host immune responses against bacterial and viral pathogens. As shown in Figs. 5 and 6, the VvTNF-α expression level decreased at 24 h after co-incubation with the PAMPs. TNF-α can induce cell necrosis, apoptosis and survival (Chu, 2013). Therefore, we speculate that the reduction of TNF-α mRNA is to protect PBLs from damage.
3.6 Protein 3D modeling analysis and protein docking simulation of VvTNF-α predict its potential functionThe secondary structure of TNF-α was predicted with Phyre2 tools (Fig. 7a). Initially, the 3D structure of VvTNF-α was predicted by SWISS-MODEL Workspace. However, the quality index, such as QMEANscore4 was -0.69, indicating that this tool was unsuitable for the 3D structure prediction in this study. Thus, we used Phyre2 to construct the 3D model (Fig. 7b) and selected d2tnfa as the template. The coverage of the protein could reach 59%, which is larger than that in SWISS-MODEL, and the confidence level could reach 100.0%. The PDBsum Generate software was used to illustrate the Ramachandran figure and predict the stability of the model (Fig. 7c). There were only a few amino acid residues in the forbidden zone. Thus, the predicted 3D structure of the VvTNF-α protein has good quality.
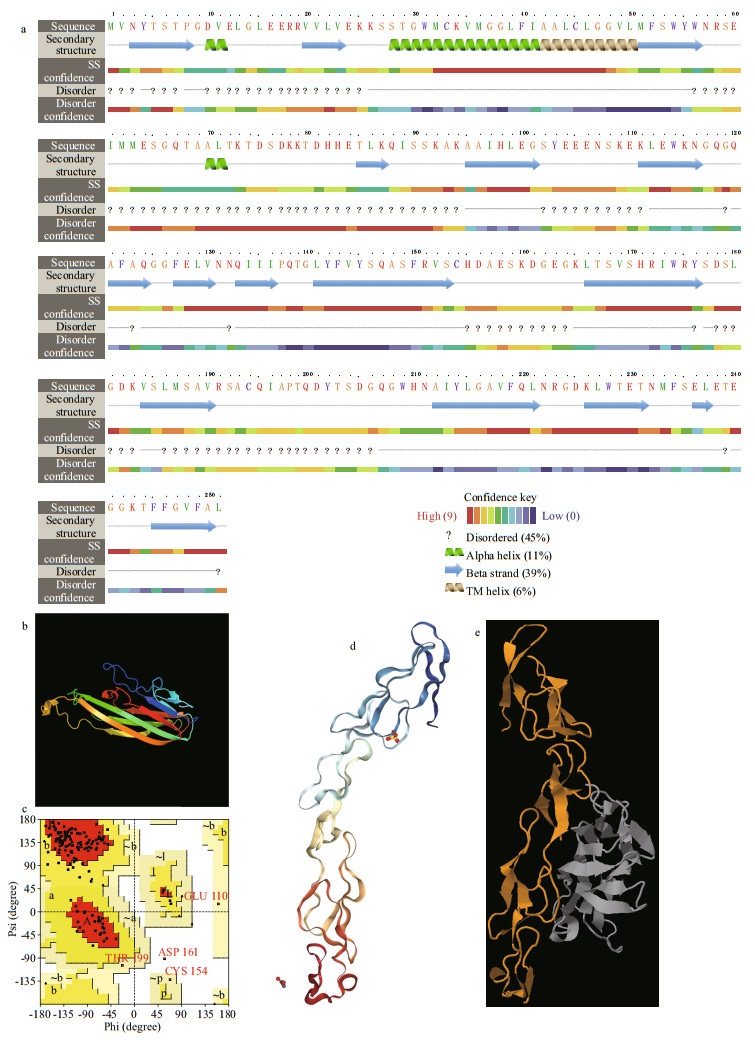
|
| Fig.7 The secondary structure, 3D structure, and protein docking simulation of the VvTNF-α The prediction was carried out by Phyre2. a. the secondary structure of VvTNF-α; b. the 3D model of VvTNF-α; c. the Ramachandran Pattern of VvTNF-α; d. potential receptor of the amyloid precursor protein (APP); e. protein docking simulation between VvTNF-α (grey) and APP (yellow) which belongs to human TNFR superfamily. |
The interaction of TNF-α and its receptor (TNFR) is the basis of the function of TNF-α (Locksley et al., 2001; Praveen et al., 2006; Xiao et al., 2007; Li and Zhang, 2016). Thus, the interaction between the predicted VvTNF-α and a potential receptor of the amyloid precursor protein (APP) (Fig. 7d), which belongs to the human TNFR super family, was tested. As shown in Fig. 7e, VvTNF-α can interact with APP through the predicted receptor binding sites.
The above results show that the 3D structure of VvTNF-α is similar to those of the other vertebrate TNF-α and imply that VvTNF-α may also perform its function by interacting with its conserved receptor binding domains.
4 CONCLUSIONWe identified the mRNA sequence of VvTNF-α and characterized its signature. The two conserved cysteine residues, TM sequence, Thr-Leu motif, and TNF family signature of this gene, as well as the TArich motifs of its proteins related to inflammatory responses were similar to the TNF-α of mammals and other reported fish. The VvTNF-α expression patterns strongly imply that it has some universal immune functions and may perform certain novel actions during the metamorphic stages. The VvTNF-α could significantly and rapidly respond to PAMPs (LPS or poly(I:C)), which indicates that this gene actively participates in host immune responses against bacterial and viral pathogens. In addition, we used the 3D structure prediction and protein docking analysis to find that the VvTNF-α function relies on the interaction with its receptors.
5 DATA AVAILABILITY STATEMENTThe sequence of TNF-α gene in spotted halibut is available from GenBank under the accession number KY038170. Additional supporting data can acquire from the corresponding author upon reasonable request.
6 CONFLICT OF INTEREST STATEMENTWe declare no conflict of interest.
Acton R D, Dahlberg P S, Uknis M E, Klaerner H G, Fink G S, Norman J G, Dunn D L. 1996. Differential sensitivity to Escherichia coli infection in mice lacking tumor necrosis factor p55 or interleukin-1 p80 receptors. Archives of Surgery, 131(11): 1 216-1 221.
DOI:10.1001/archsurg.1996.01430230098017 |
Aust G, Heuer M, Laue S, Lehmann I, Hofmann A, Heldin N E, Scherbaum W A. 1996. Expression of tumour necrosis factor-alpha (TNF-α) mRNA and protein in pathological thyroid tissue and carcinoma cell lines. Clinical and Experimental Immunology, 105(1): 148-154.
DOI:10.1046/j.1365-2249.1996.d01-726.x |
Beg A A, Baltimore D. 1996. An essential role for NF-κB in preventing TNF-α-induced cell death. Science, 274(5288): 782-784.
DOI:10.1126/science.274.5288.782 |
Blobel C P. 1997. Metalloprotease-disintegrins:links to cell adhesion and cleavage of TNFα and Notch. Cell, 90(4): 589-592.
DOI:10.1016/S0092-8674(00)80519-X |
Campinho M A, Silva N, Sweeney G E, Power D M. 2007. Molecular, cellular and histological changes in skin from a larval to an adult phenotype during bony fish metamorphosis. Cell and Tissue Research, 327(2): 267-284.
|
Camussi G, Albano E, Tetta C, Bussolino F. 1991. The molecular action of tumor necrosis factor-α. European Journal of Biochemistry, 202(1): 3-14.
|
Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. 1986. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proceedings of the National Academy of Sciences of the United States of America, 83(6): 1 670-1 674.
DOI:10.1073/pnas.83.6.1670 |
Chang M X, Nie P, Xie H X, Wang G L, Gao Y. 2006. Characterization and expression analysis of TNF-related apoptosis inducing ligand (TRAIL) in grass carp Ctenopharyngodon idella. Veterinary Immunology and Immunopathology, 110(1-2): 51-63.
DOI:10.1016/j.vetimm.2005.09.003 |
Chu W. 2013. Tumor necrosis factor. Cancer Letters, 328(2): 222-225.
DOI:10.1016/j.canlet.2012.10.014 |
Covello J M, Bird S, Morrison R N, Battaglene S C, Secombes C J, Nowak B F. 2009. Cloning and expression analysis of three striped trumpeter (Latris lineata) pro-inflammatory cytokines, TNF-α, IL-1β and IL-8, in response to infection by the ectoparasitic, Chondracanthus goldsmidi. Fish & Shellfish Immunology, 26(5): 773-786.
DOI:10.1016/j.fsi.2009.03.012 |
De Jesus E G, Hirano T, Inui Y. 1991. Changes in cortisol and thyroid hormone concentrations during early development and metamorphosis in the Japanese flounder, Paralichthys olivaceus. General and Comparative Endocrinology, 82(3): 369-376.
DOI:10.1016/0016-6480(91)90312-T |
Ellis A E. 2001. Innate host defense mechanisms of fish against viruses and bacteria. Developmental & Comparative Immunology, 25(8-9): 827-839.
|
Forlenza M, Magez S, Scharsack J P, Westphal A, Savelkoul H F J, Wiegertjes G F. 2009. Receptor-mediated and lectinlike activities of carp (Cyprinus carpio) TNF-α. Journal of Immunology, 183(8): 5 319-5 332.
DOI:10.4049/jimmunol.0901780 |
García-Castillo J, Chaves-Pozo E, Olivares P, Pelegrín P, Meseguer J, Mulero V. 2004. The tumor necrosis factor α of the bony fish seabream exhibits the in vivo proinflammatory and proliferative activities of its mammalian counterparts, yet it functions in a speciesspecific manner. Cellular and Molecular Life Sciences CMLS, 61(11): 1 331-1 340.
DOI:10.1007/s00018-004-4068-1 |
García-Castillo J, Pelegrín P, Mulero V, Meseguer J. 2002. Molecular cloning and expression analysis of tumor necrosis factor α from a marine fish reveal its constitutive expression and ubiquitous nature. Immunogenetics, 54(3): 200-207.
DOI:10.1007/s00251-002-0451-y |
Grayfer L, Walsh J G, Belosevic M. 2008. Characterization and functional analysis of goldfish (Carassius auratus L. )tumor necrosis factor-alpha. Developmental & Comparative Immunology, 32(5): 532-543.
DOI:10.1016/j.dci.2007.09.009 |
Han J, Brown T, Beutler B. 1990. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. Journal of Experimental Medicine, 171(2): 465-475.
DOI:10.1084/jem.171.2.465 |
Hawiger J. 2001. Innate immunity and inflammation:a transcriptional paradigm. Immunologic Research, 23(2-3): 99-109.
DOI:10.1385/IR:23:2-3:099 |
Hirono I, Nam B H, Kurobe T, Aoki T. 2000. Molecular cloning, characterization, and expression of TNF cDNA and gene from Japanese flounder Paralychthys olivaceus. Journal of Immunology, 165(8): 4 423-4 427.
DOI:10.4049/jimmunol.165.8.4423 |
Hong S, Li R G, Xu Q Q, Secombes C J, Wang T H. 2013. Two types of TNF-α Exist in teleost fish:phylogeny, expression, and bioactivity analysis of Type-Ⅱ TNF-α3 in rainbow trout Oncorhynchus mykiss. Journal of Immunology, 191(12): 5 959-5 972.
DOI:10.4049/jimmunol.1301584 |
Hwang S D, Shim S H, Kwon M, Chae Y S, Shim W J, Jung J H, Kim J W, Park C I. 2014. Molecular cloning and expression analysis of two lipopolysaccharide-induced TNF-α factors (LITAFs) from rock bream, Oplegnathus fasciatus. Fish & Shellfish Immunology, 36(2): 467-474.
DOI:10.1016/j.fsi.2013.12.023 |
Idriss H T, Naismith J H. 2000. TNFα and the TNF receptor superfamily:structure-function relationship(s). Microscopy Research and Technique, 50(3): 184-195.
DOI:10.1002/1097-0029(20000801)50:3<184::AID-JEMT2>3.0.CO;2-H |
Inui Y, Miwa S. 1985. Thyroid hormone induces metamorphosis of flounder larvae. General and Comparative Endocrinology, 60(3): 450-454.
DOI:10.1016/0016-6480(85)90080-2 |
Jang C H, Choi J H, Byun M S, Jue D M. 2006. Chloroquine inhibits production of TNF-α, IL-1β and IL-6 from lipopolysaccharide-stimulated human monocytes/macrophages by different modes. Rheumatology, 45(6): 703-710.
DOI:10.1093/rheumatology/kei282 |
Kalashnikova S A, Goryachev A N, Novochadov V V, Shchyogolev A I. 2009. Thyroid modulation of TNFdependent apoptosis and formation of chronic liver disease in endogenous intoxication in rats. Bulletin of Experimental Biology and Medicine, 147(2): 240-244.
DOI:10.1007/s10517-009-0484-4 |
Kim B S, Nam B H, Kim J W, Park H J, Song J H, Park C I. 2011. Molecular characterisation and expression analysis of a fish-egg lectin in rock bream, and its response to bacterial or viral infection. Fish & Shellfish Immunology, 31(6): 1 201-1 207.
|
Kiss-Toth E, Harlock E, Lath D, Quertermous T, Wilkinson J M. 2013. A TNF Variant that associates with susceptibility to musculoskeletal disease modulates thyroid hormone receptor binding to control promoter activation. PLoS One, 8(9): e76034.
DOI:10.1371/journal.pone.0076034 |
Klaren P H M, Wunderink Y S, Yúfera M, Mancera J M, Flik G. 2008. The thyroid gland and thyroid hormones in Senegalese sole (Solea senegalensis) during early development and metamorphosis. General and Comparative Endocrinology, 155(3): 686-694.
DOI:10.1016/j.ygcen.2007.09.014 |
Li H J, Fan J F, Liu S X, Yang Q, Mu G Q, He C B. 2012. Characterization of a myostatin gene (MSTN1) from spotted halibut (Verasper variegatus) and association between its promoter polymorphism and individual growth performance. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 161(4): 315-322.
DOI:10.1016/j.cbpb.2011.12.008 |
Li H J, Jiang L X, Han J B, Su H, Yang Q, He C B. 2011. Major histocompatibility complex class IIA and IIB genes of the spotted halibut Verasper variegatus:genomic structure, molecular polymorphism, and expression analysis. Fish Physiology and Biochemistry, 37(4): 767-780.
DOI:10.1007/s10695-011-9476-1 |
Li M F, Zhang J. 2016. CsTNF1, a teleost tumor necrosis factor that promotes antibacterial and antiviral immune defense in a manner that depends on the conserved receptor binding site. Developmental & Comparative Immunology, 55: 65-75.
|
Liu Y W, Chan W K. 2002. Thyroid hormones are important for embryonic to larval transitory phase in zebrafish. Differentiation, 70(1): 36-45.
DOI:10.1046/j.1432-0436.2002.700104.x |
Locksley R M, Killeen N, Lenardo M J. 2001. The TNF and TNF receptor superfamilies:integrating mammalian biology. Cell, 104(4): 487-501.
DOI:10.1016/S0092-8674(01)00237-9 |
Loetscher H, Steinmetz M, Lesslauer W. 1991. Tumor necrosis factor:receptors and inhibitors. Cancer Cells, 3(6): 221-226.
|
Marchand O, Duffraisse M, Triqueneaux G, Safi R, Laudet V. 2004. Molecular cloning and developmental expression patterns of thyroid hormone receptors and T3 target genes in the turbot (Scophtalmus maximus) during postembryonic development. General and Comparative Endocrinology, 135(3): 345-357.
DOI:10.1016/j.ygcen.2003.10.012 |
Marchler-Bauer A, Anderson J B, Chitsaz F, Derbyshire M K, DeWeese-Scott C, Fong J H, Geer L Y, Geer R C, Gonzales N R, Gwadz M, He S Q, Hurwitz D I, Jackson J D, Ke Z X, Lanczycki C J, Liebert C A, Liu C L, Lu F, Lu S N, Marchler G H, Mullokandov M, Song J S, Tasneem A, Thanki N, Yamashita R A, Zhang D C, Zhang N G, Bryant S H. 2009. CDD:specific functional annotation with the Conserved Domain Database. Nucleic Acids Research, 37(S1): 205-210.
|
Marchler-Bauer A, Bryant S H. 2004. CD-Search:protein domain annotations on the fly. Nucleic Acids Research, 32(S2): 327-331.
|
Marchler-Bauer A, Derbyshire M K, Gonzales N R, Lu S N, Chitsaz F, Geer L Y, Geer R C, He J N, Gwadz M, Hurwitz D I, Lanczycki C J, Lu F, Marchler G H, Song J S, Thanki N, Wang Z X, Yamashita R A, Zhang D C, Zheng C J, Bryant S H. 2015. CDD:NCBI's conserved domain database. Nucleic Acids Research, 43(D1): D222-D226.
DOI:10.1093/nar/gku1221 |
Marchler-Bauer A, Lu S N, Anderson J B, Chitsaz F, Derbyshire M K, DeWeese-Scott C, Fong J H, Geer L Y, Geer R C, Gonzales N R, Gwadz M, Hurwitz D I, Jackson J D, Ke Z X, Lanczycki C J, Lu F, Marchler G H, Mullokandov M, Omelchenko M V, Robertson C L, Song J S, Thanki N, Yamashita R A, Zhang D C, Zhang N Q, Zheng C J, Bryant S H. 2011. CDD:a conserved domain database for the functional annotation of proteins. Nucleic Acids Research, 39(D1): D225-D229.
|
McGeehan G M, Becherer J D, Bast R C Jr, Boyer C M, Champion B, Connolly K M, Conway J G, Furdon P, Karp S, Kidao S, McElroy A B, Nichols J, Pryzwansky K M, Schoenen F, Sekut L, Truesdale A, Verghese M, Warner J, Ways J P. 1994. Regulation of tumour necrosis factor-α processing by a metalloproteinase inhibitor. Nature, 370(6490): 558-561.
DOI:10.1038/370558a0 |
Nascimento D S, Pereira P J B, Reis M I R, Do Vale A, Zou J, Silva M T, Secombes C J, Dos Santos N M S. 2007. Molecular cloning and expression analysis of sea bass(Dicentrarchus labrax L.) tumor necrosis factor-α(TNF-α). Fish & Shellfish Immunology, 23(3): 701-710.
DOI:10.1016/j.fsi.2007.02.003 |
Olleros M L, Guler R, Corazza N, Vesin D, Eugster H P, Marchal G, Chavarot P, Mueller C, Garcia I. 2002. Transmembrane TNF induces an efficient cell-mediated immunity and resistance to Mycobacterium bovis bacillus Calmette-Guerin infection in the absence of secreted TNF and lymphotoxin-α. Journal of Immunology, 168(7): 3 394-3 401.
DOI:10.4049/jimmunol.168.7.3394 |
Ordás M C, Costa M M, Roca F J, López-Castejón G, Mulero V, Meseguer J, Figueras A, Novoa B. 2007. Turbot TNFα gene:molecular characterization and biological activity of the recombinant protein. Molecular Immunology, 44(4): 389-400.
DOI:10.1016/j.molimm.2006.02.028 |
Pasparakis M, Alexopoulou L, Episkopou V, Kollias G. 1996. Immune and inflammatory responses in TNF α-deficient mice:a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. Journal of Experimental Medicine, 184(4): 1 397-1 411.
DOI:10.1084/jem.184.4.1397 |
Pennica D, Nedwin G E, Hayflick J S, Seeburg P H, Derynck R, Palladino M A, Kohr W J, Aggarwal B B, Goeddel D V. 1984. Human tumour necrosis factor:precursor structure, expression and homology to lymphotoxin. Nature, 312(5996): 724-729.
DOI:10.1038/312724a0 |
Power D M, Llewellyn L, Faustino M, Nowell M A, Björnsson B T, Einarsdottir I E, Canario A V M, Sweeney G E. 2001. Thyroid hormones in growth and development of fish. Comparative Biochemistry and Physiology Part C:Toxicology & Pharmacology, 130(4): 447-459.
|
Praveen K, Evans D L, Jaso-Friedmann L. 2006. Constitutive expression of tumor necrosis factor-alpha in cytotoxic cells of teleosts and its role in regulation of cell-mediated cytotoxicity. Molecular Immunology, 43(3): 279-291.
DOI:10.1016/j.molimm.2005.01.012 |
Qi P Z, Xie C X, Guo B Y, Wu C W. 2016. Dissecting the role of transforming growth factor-β1 in topmouth culter immunobiological activity:a fundamental functional analysis. Scientific Reports, 6: 27179.
DOI:10.1038/srep27179 |
Rink L, Kirchner H. 1996. Recent progress in the tumor necrosis factor-α field. International Archives of Allergy and Immunology, 111(3): 199-209.
DOI:10.1159/000237369 |
Roca F J, Mulero I, López-Muñoz A, Sepulcre M P, Renshaw S A, Meseguer J, Mulero V. 2008. Evolution of the inflammatory response in vertebrates:fish TNF-α is a powerful activator of endothelial cells but hardly activates phagocytes. Journal of Immunology, 181(7): 5 071-5 081.
DOI:10.4049/jimmunol.181.7.5071 |
Sachs A B. 1993. Messenger RNA degradation in eukaryotes. Cell, 74(3): 413-421.
DOI:10.1016/0092-8674(93)80043-E |
Saeij J P J, Stet R J M, De Vries B J, Van Muiswinkel W B, Wiegertjes G F. 2003. Molecular and functional characterization of carp TNF:a link between TNF polymorphism and trypanotolerance. Developmental and Comparative Immunology, 27(1): 29-41.
DOI:10.1016/S0145-305X(02)00064-2 |
Saunders B M, Tran S, Ruuls S, Sedgwick J D, Briscoe H, Britton W J. 2005. Transmembrane TNF is sufficient to initiate cell migration and granuloma formation and provide acute, but not long-term, control ofMycobacterium tuberculosis infection. Journal of Immunology, 174(8): 4 852-4 859.
DOI:10.4049/jimmunol.174.8.4852 |
Savan R, Sakai M. 2004. Presence of multiple isoforms of TNF alpha in carp (Cyprinus carpio L.):genomic and expression analysis. Fish & Shellfish Immunology, 17(1): 87-94.
|
Sherry B, Cerami A. 1988. Cachectin/tumor necrosis factor exerts endocrine, paracrine, and autocrine control of inflammatory responses. Journal of Cell Biology, 107(4): 1 269-1 277.
DOI:10.1083/jcb.107.4.1269 |
Singer S J. 1990. The structure and insertion of integral proteins in membranes. Annual Review of Cell Biology, 6: 247-296.
DOI:10.1146/annurev.cb.06.110190.001335 |
Teles M, Mackenzie S, Boltaña S, Callol A, Tort L. 2011. Gene expression and TNF-alpha secretion profile in rainbow trout macrophages following exposures to copper and bacterial lipopolysaccharide. Fish & Shellfish Immunology, 30(1): 340-346.
|
Uysal K T, Wiesbrock S M, Marino M W, Hotamisligil G S. 1997. Protection from obesity-induced insulin resistance in mice lacking TNF-α function. Nature, 389(6651): 610-614.
DOI:10.1038/39335 |
Wang Y S, Bu L Z, Yang L L, Li H Y, Zhang S C. 2016. Identification and functional characterization of fish-egg lectin in zebrafish. Fish & Shellfish Immunology, 52: 23-30.
|
Wride M A, Sanders E J. 1995. Potential roles for tumour necrosis factor α during embryonic development. Anatomy and Embryology, 191(1): 1-10.
DOI:10.1007/bf00215292 |
Xiao J, Zhou Z C, Chen C, Huo W L, Yin Z X, Weng S P, Chan S M, Yu X Q, He J G. 2007. Tumor necrosis factor-alpha gene from mandarin fish, Siniperca chuatsi:Molecular cloning, cytotoxicity analysis and expression profile. Molecular Immunology, 44(14): 3 615-3 622.
DOI:10.1016/j.molimm.2007.03.016 |
Xu Y J, Liu X Z, Liao M J, Wang H P, Wang Q Y. 2012. Molecular cloning and differential expression of three GnRH genes during ovarian maturation of spotted halibut, Verasper variegatus. Journal of Experimental Zoology, Part A, Ecological Genetics and Physiology, 317(7): 434-446.
DOI:10.1002/jez.1736 |
Yamano K, Araki K, Sekikawa K, Inui Y. 1994. Cloning of thyroid hormone receptor genes expressed in metamorphosing flounder. Developmental Genetics, 15(4): 378-382.
DOI:10.1002/dvg.1020150409 |
Yamano K, Miwa S. 1998. Differential gene expression of thyroid hormone receptor α and β in fish development. General and Comparative Endocrinology, 109(1): 75-85.
DOI:10.1006/gcen.1997.7011 |
Zhang A Y, Chen D Y, Wei H, Du L Y, Zhao T Q, Wang X Y, Zhou H. 2012. Functional characterization of TNF-α in grass carp head kidney leukocytes:Induction and involvement in the regulation of NF-κB signaling. Fish & Shellfish Immunology, 33(5): 1 123-1 132.
|
Zhou Z X, Zhang B C, Sun L. 2014. Poly(I:C) induces antiviral immune responses in Japanese flounder (Paralichthys olivaceus) that require TLR3 and MDA5 and is negatively regulated by Myd88. PLoS One, 9(11): e112918.
DOI:10.1371/journal.pone.0112918 |
Zou J, Secombes C J, Long S, Miller N, Clem L W, Chinchar V G. 2003. Molecular identification and expression analysis of tumor necrosis factor in channel catfish(Ictalurus punctatus). Developmental & Comparative Immunology, 27(10): 845-858.
|
Zou J, Wang T, Hirono I, Aoki T, Inagawa H, Honda T, Soma G, Ototake M, Nakanishi T, Ellis A E, Secombes C J. 2002. Differential expression of two tumor necrosis factor genes in rainbow trout, Oncorhynchus mykiss. Developmental & Comparative Immunology, 26(2): 161-172.
|
 2020, Vol. 38
2020, Vol. 38