Institute of Oceanology, Chinese Academy of Sciences
Article Information
- PARK YeonJung, LEE Mi Nan, KIM Eun-Mi, PARK JungYoun, NOH JaeKoo, CHOI Tae-Jin, KANG Jung-Ha
- Development and characterization of novel polymorphic microsatellite markers for the Korean freshwater snail Semisulcospira coreana and cross-species amplification using next-generation sequencing
- Journal of Oceanology and Limnology, 38(2): 503-508
- http://dx.doi.org/10.1007/s00343-019-9058-0
Article History
- Received Mar. 12, 2019
- accepted in principle Apr. 11, 2019
- accepted for publication Jun. 4, 2019
2 Department of Microbiology, Pukyoung National University, Busan 48513, Republic of Korea
Korean freshwater snails of the genus Semisulcospira are a prominent freshwater gastropod resource across East Asia, including in Korea, Japan, southeastern China, and Taiwan, China (Davis, 1969). Freshwater snails have been used as healthy food sources in Korea and play important roles as biological indicators in the environmental monitoring of freshwater systems (Lee and Lim, 2005). Currently, the snail population is rapidly declining in Korea due to increasing human consumption, overharvesting, habitat degradation, and water pollution caused by insecticides and the restructuring of riverbeds (Kim et al., 2010). In order to recover the decreasing resources, artificial seed production was being promoted. In addition, a large number of foreign snail species have been imported to recover the declining population (Moon et al., 2015). Many species belonging to this genus, including S. coreana, S. tegulata, S. libertina, S. forticosta, and S. gottschei, are produced in different regions. Especially, the Korean endemic species is S. coreana, which is distributed in rivers of the Midwest region, such as the Geum River, Seomjin River, and Yeongsan River in Korea (Kim et al., 2012). Of the genus, S. coreana and S. libertina that recognized as the major economic resource on the inland waters are principal culture species. Despite of its ecological importance, there have been a few phylogenetic, phenotypic, and genetic analyses (Urabe, 2000; Zeng et al., 2015); no population-level study using microsatellite markers has been carried out to clarify genetic diversity and population structure in the genus Semisulcospira.
Among the currently available DNA-based techniques, microsatellite DNA markers, also named simple sequence repeats (SSRs), are useful tools to assess the genetic variance and structure of populations because of their several desirable features such as variability, codominance, and high mutation rates (Féral, 2002). They have become the most widely useful DNA technology in many fields of biology, including genome mapping, determining pedigree, population genetics, biological resource conservation, and forensic studies (Knapik et al., 1998; Luikart et al., 2003). The traditional cloning methods used to develop microsatellites involved significant trial and error, required knowledge of the flanking region DNA sequence, and were time-consuming, cost prohibitive, and low throughput (Queller et al., 1993). At present, these problems have been partly resolved with the advent of next-generation sequencing (NGS) technologies that are both cost- and time-effective, as they can manufacture millions of base pairs of short fragment reads in a single run (Moges et al., 2016). Furthermore, when primers newly developed for one species can be used for broad taxonomic groups is increased the cost- and time-effectiveness of microsatellites. The cross-species amplification of microsatellites has successfully been applied to several marine species (Greenley et al., 2012).
In the present study, we developed the first 18 novel polymorphic microsatellite markers for S. coreana and tested cross-species amplification in four additional Semisulcospira spp. These loci will assist in obtaining genetic information for resource management of the genus Semisulcospira throughout the Korean drainage area.
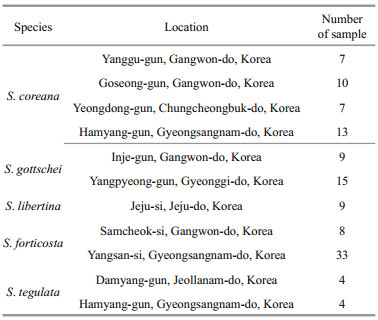
2 MATERIAL AND METHOD 2.1 Sample collection and DNA extractionThirty-seven individuals of S. coreana were obtained from the Inland Fisheries Research Institute, National Institute of Fisheries Science (NIFS) (Table 1). Foot muscles from the samples were maintained in 100% ethanol before being transported to the laboratory. Genomic DNA extraction procedure was conducted using the MagExtractor with an MFX- 6100 automated DNA extraction system (Toyobo, Osaka, Japan) under the manufacturer's instructions. The concentration and quality of extracted genomic DNA was estimated using a spectrophotometer NanoDrop ND-1000 (Thermo Fisher Scientific, Barrington, IL, USA). For the cross-species transferability test to other Semisulcospira species including S. tegulata, S. libertina, S. forticosta, and S. gottschei, DNA extraction was performed by the same method from ethanol-fixed tissue from four related Semisulcospira spp. obtained from NIFS in Ulgin, Korea.
The NGS library was generated from ~10 μg genomic DNA and sequenced on a GS-FLX-454 pyrosequencing system (454 Life Sciences, Branford, CT, USA) at the NICEM (National Instrumentation Center for Environmental Management of Seoul National University). The obtained sequence reads were trimmed to 96% minimum overlap identity using Newbler 2.6 (Roche Diagnostics, Mannheim, Germany). To search for SSRs in the genomic sequence, the dinucleotide and trinucleotide repeats of more than seven iterations were screened using the Perl program SSR_finder.pl (Tóth et al., 2000). Primer pairs complementary to the sequence flanking the repeat element were designed using Primer 3 software (Untergasser et al., 2012). The optimal size for primer was set to a range as 18–26 bases and the optimal annealing temperature was set at 58℃. The optimal product size was set to 130–400 bp and the remaining parameters were kept at default settings.
2.3 DNA amplification and genotypingThe performances of the newly developed primer sets were tested for optimum concentration for PCR amplification using DNA from eight individuals of S. coreana. The electrophoresis of PCR products were operated on a 1.5% agarose gel and 40 primer sets produced PCR products of 100–300 bp in length. Thirty of these primer sets were labeled and used to amplify DNA from the eight individuals. The forward primer was labeled at 5′ end with the fluorescent dyes 6-FAM, TAMRA, and HEX (Applied Biosystems, Foster City, CA, USA). PCR amplifications were performed in 10-μL total volumes that included 0.4 μmol/L of the forward and reverse primer, 0.2 mmol/L dNTPs, 1× PCR buffer, 0.25 U Ex Taq DNA polymerase (TaKaRa Biomedical, Inc., Shiga, Japan), and approximately 100 ng template DNA under the following conditions: pre-denaturation at 95℃ for 10 min, followed by 35 cycles of 45 s at 94℃, 45 s of 58℃, and 45 s at 72℃ with a finishing of 5 min at 72℃ using an ABI 2720 Thermocycler (Applied Biosystems, USA). The PCR products were detected on a 1.5% agarose gel. Fragment analysis was run on a 3730xl DNA Analyzer (Applied Biosystems, USA) using GeneScan 400HD ROX dye as the internal size standard (Life Technologies, Carlsbad, CA, USA) and analyzed using GeneMapper 5.0 (Applied Biosystems). Primer sets that generated numerous signal peaks or amplified monomorphic microsatellite loci were excluded.
2.4 Transferability analysisTo test the cross-species transferability of the SSR loci, we used all 18 polymorphic sites of S. coreana on samples of four related taxa. Transferability tests were performed using the abovementioned PCR conditions. Amplification products were monitored on 1.5% agarose gels, and qualitatively graded on the presence or absence of the band.
2.5 Data analysisNumbers of alleles, effective numbers of alleles, expected and observed heterozygosities were assessed using Arlequin 3.0 (Excoffier and Lischer, 2010). The significance level (P-value) for Hardy-Weinberg equilibrium (HWE) was evaluated using Genepop 4.0 (Rousset, 2008). Polymorphic information content (PIC) was assessed statistically using Cervus 3.0 program (Kalinowski et al., 2007). The presence of null alleles and potential genotyping errors was calculated statistically using MicroChecker 2.2.3 (Van Oosterhout et al., 2004).
3 RESULT AND DISCUSSIONNGS generated 17 719 000 reads of a total of 8 894 938 000 bp for S. coreana. The total number of contigs was 132 872 and the number of singletons was 1 754 529. The total number of reads after trimming low-quality sequences was 9 967 563. A total of 3 565 825 918 regions were found to contain from nucleotide repeats for more than four iterations. The amplicon sizes varied from 90 to 320 bp. Of them 41%, 27%, and 32% were di-, tri-, and tetranucleotides, respectively. CA repeats were the most common dinucleotide repeats, TTG repeats were the most common trinucleotide repeats, and TGTG repeats were the most common tetra-nucleotide repeats.
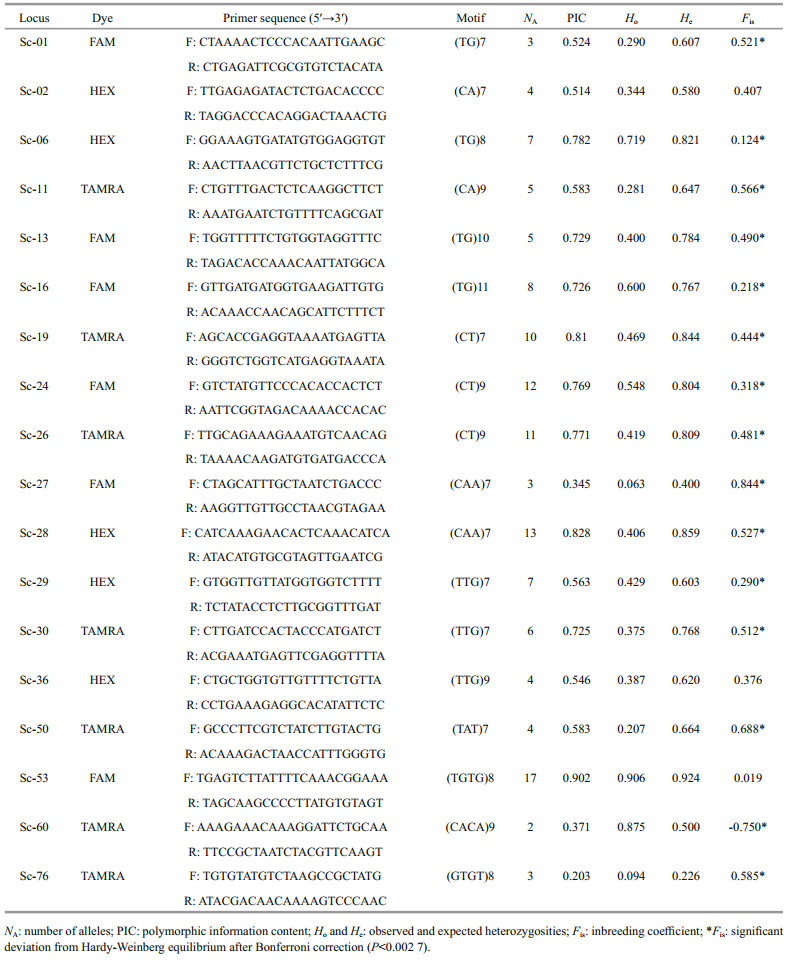
All of 78 SSRs with high copy numbers of all repeat motifs for the amplification and assessment of polymorphisms were synthesized to measure amplification efficiency and extent of polymorphism. Among them, a total of 18 primer pairs (23%) showing clear amplification of polymorphic loci were applied for PCR analysis and genotyping. The primer details and fluorescent labeling information are shown in Table 2.
In the present, microsatellite developed in most marine resource, was mostly consisted as dinucleotide repeats. However, tri- and tetra-nucleotides motif have the advantages like highly polymorphic and stable than di-nucleotide repeats (Lindqvist et al., 1996). We developed 18 novel polymorphic microsatellite markers consisted of all of di-, tri-, and tetranucleotide motifs. The allele number per locus resulted from 2 to 17, with a mean of 6.9. The PIC for each locus ranged from 0.203 to 0.902, with a mean of 0.626. Botstein et al. (1980) described that a PIC value above 0.5 has sufficient discrimination within a population. In addition, comparisons of PIC values can present an estimate of the power of the marker. Fifteen of the 18 loci were supposed to be highly informative (PIC>0.5). Another two (Sc-27 and Sc- 60) were indicated reasonably informative (0.25 < PIC < 0.5), and one (Sc-76) was only slightly informative (PIC < 0.25). This suggests that the set of microsatellites generated in the present study shows considerable potential for analyzing genetic polymorphisms.
The observed and expected heterozygosities of the loci resulted from 0.063 to 0.924 and 0.226 to 0.924, respectively. The inbreeding coefficient (Fis) ranged from -0.750 to 0.844. Null alleles were detected at most of the loci, except for Sc06, Sc53, and Sc60, which showed deviation from HWE after Bonferroni correction (P < 0.002 7) with expected heterozygosity (He) values greater than observed heterozygosity (Ho) values. The significant deficits of heterozygosity are associated with the existence of null alleles (Selkoe and Toonen, 2006), which are common in freshwater mollusca (Gu at al., 2012). This is also demonstrated by the significant positive Fis values at most of the loci, except for Sc-60, indicating that the excess of homozygotes was likely due to the existence of null alleles or stuttering at these loci, as suggested by the Micro-Checker results. This is also caused by inbreeding within a population in invertebrates due to the poor mobility of freshwater snails (Gu et al., 2015).
Cross-species amplification of the 18 polymorphic loci was conducted within four species of Semisulcospira, S. forticosta, S. gottschei, S. libertina, and S. tegulata (Table 3). As shown in Table 3, three polymorphic loci (Sc-50, Sc-60, and Sc-76) were not amplified by any species tested. These three loci could be used as identification of S. coreana. Loci Sc-24 showed polymorphic amplification in all species tested except S. libertina. The transferability test of these microsatellite markers for the four species showed a very high transferability (80%–85%). The transferability of SSR markers in different snail species is affected by the nucleotide sequence similarity in the SSR marker primer sites among related species (Barbará et al., 2007). The crossspecies transferability of SSR primers in the snails was high, indicating that there is high conservation of markers between Semisulcospira spp.

|
We developed 18 new microsatellite markers for S. coreana using 454 GS-FLX NGS. This is the first set of polymorphic microsatellite markers designed for S. coreana and related species. These microsatellite markers will potentially be useful for genetic diversity and population genetic structure analyses of snails throughout the Korean drainage area.
4 CONCLUSIONKorean freshwater snails of the genus Semisulcospira are economically and ecologically important inland resource. To study population genetic diversity of this genus, the development of microsatellite marker is necessary. The 18 novel polymorphic microsatellite loci from Semisulcospira coreana were isolated using 454 GS-FLX titanium sequencing. In addition, the transferability of all of the loci was checked on S. forticosta, S. gottschei, S. tegulata, and S. libertina. These novel microsatellite markers will aid freshwater snail resource investigations and population genetic conservation.
5 DATA AVAILABILITY STATEMENTAll data generated and/or analyzed during this study are included in this article, or are available upon the request from the readers.
6 COMPLIANCE WITH ETHICAL STANDARDThe authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
Barbará T, Palma-Silva C, Paggi G M, Bered F, Fay M F, Lexer C. 2007. Cross-species transfer of nuclear microsatellite markers:potential and limitations. Mol. Ecol., 16(18): 3759-3767.
DOI:10.1111/j.1365-294X.2007.03439.x |
Davis G M. 1969. A taxonomic study of some species of Semisulcospira in Japan (Mesogastropoda:Pleuroceridae). Malacologia, 7(2-3): 211-294.
|
Botstein D, White R L, Skolnick M, Davis R W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum.Genet., 32: 314-331.
|
Excoffier L, Lischer H E. 2010. Arlequin suite Ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour., 10(3): 564-567.
DOI:10.1111/j.1755-0998.2010.02847.x |
Féral J P. 2002. How useful are the genetic markers in attempts to understand and manage marine biodiversity? J. Exp.Mar. Biol. Ecol., 268(2): 121-145.
DOI:10.1016/S0022-0981(01)00382-3 |
Greenley A P, Muguia-Vega A, Saenz-Arroyo A, Micheli F. 2012. New tetranucleotide microsatellite loci in pink abalone (Haliotis corrugata) isolated via 454 pyrosequencing. Conservat. Genet. Resour., 4(2): 265-268.
DOI:10.1007/s12686-011-9521-5 |
Gu Q H, Cheng Q Q, Li X J, Zhou C J. 2015. Novel polymorphic microsatellite markers for Bellamya and their application in population genetics of three species. Genet. Mol. Res., 14(4): 15201-15212.
DOI:10.4238/2015.November.25.8 |
Gu Q H, Xiong B X, Zhu Y T, Wang Q, Shi P S. 2012. Isolation and characterization of polymorphic microsatellites loci in the freshwater snail Bellamya aeruginosa (Mollusca, Gastropoda). Conservat. Genet. Resour., 4(2): 387-390.
DOI:10.1007/s12686-011-9556-7 |
Kalinowski S T, Taper M L, Marshall T C. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol., 16(5): 1099-1106.
DOI:10.1111/j.1365-294X.2007.03089.x |
Kim D H, Bang I C, Lee W O, Baek J M. 2012. Reproductive ecology of the freshwater Melania snail, Semisulcospira coreana (V.Martens) in Bukhan River. Korean J.Malacol., 28(2): 175-185.
DOI:10.9710/kjm.2012.28.2.175 |
Kim W J, Kim D H, Lee J S, Bang I C, Lee W O, Jung H. 2010. Systematic relationships of Korean freshwater snails of Semisulcospira, Koreanomelania, and Koreoleptoxis (Cerithiodiea; Pleuroceridae) revealed by mitochondrial cytochrome oxidase I sequences. Korean J. Malacol., 26(4): 275-283.
|
Knapik E W, Goodman A, Ekker M, Chevrette M, Delgado J, Neuhauss S, Shimoda N, Driever W, Fishman M C, Jacob H J. 1998. A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat. Genet., 18(4): 338-343.
DOI:10.1038/ng0498-338 |
Lee S M, Lim T J. 2005. Effects of dietary protein and energy levels on growth and lipid composition of juvenile snail(Semisulcospira gottschei). J. Shellfish Res., 24(1): 99-102.
DOI:10.2983/0730-8000(2005)24[99:EODPAE]2.0.CO;2 |
Lindqvist A K, Magnusson P K, Balciuniene J, Wadelius C, Lindholm E, Alarcón-Riquelme M E, Gyllensten U B. 1996. Chromosome-specific panels of tri- and tetranucleotide microsatellite markers for multiplex fluorescent detection and automated genotyping:evaluation of their utility in pathology and forensics. Genome Res., 6(12): 1170-1176.
DOI:10.1101/gr.6.12.1170 |
Luikart G, England P R, Tallmon D, Jordan S, Taberlet P. 2003. The power and promise of population genomics:from genotyping to genome typing. Nat. Rev. Genet., 4(12): 981-994.
|
Moges A D, Admassu B, Belew D, Yesuf M, Njuguna J, Kyalo M, Ghimire S R. 2016. Development of microsatellite markers and analysis of genetic diversity and population structure of Colletotrichum gloeosporioides from Ethiopia. PLoS One, 11(3): e0151257.
DOI:10.1371/journal.pone.0151257 |
Moon S K, Kim I S, Lim C W, Yoon N Y, Jeong B Y. 2015. Proximate and fatty acid compositions of commercial domestic and imported Melania snails Semisulscospira sp. Korean J. Fish. Aquat. Sci., 48(6): 977-981.
|
Queller D C, Strassmann J E, Hughes C R. 1993. Microsatellites and kinship. Trends Ecol. Evol., 8(8): 285-288.
DOI:10.1016/0169-5347(93)90256-O |
Rousset F. 2008. GENEPOP'007:a complete reimplementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour., 8(1): 103-106.
DOI:10.1111/j.1471-8286.2007.01931.x |
Selkoe K A, Toonen R J. 2006. Microsatellites for ecologists:a practical guide to using and evaluating microsatellite markers. Ecol. Lett., 9(5): 615-629.
DOI:10.1111/j.1461-0248.2006.00889.x |
Tóth G, Gáspári Z, Jurka J. 2000. Microsatellites in different eukaryotic genomes:survey and analysis. Genome Res., 10(7): 967-981.
DOI:10.1101/gr.10.7.967 |
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth B C, Remm M, Rozen S G. 2012. Primer3-new capabilities and interfaces. Nucleic Acids Res., 40(15): e115.
DOI:10.1093/nar/gks596 |
Urabe M. 2000. Phenotypic modulation by the substratum of shell sculpture in Semisulcospira reiniana (Prosobranchia:Pleuroceridae). J. Mol. Stud., 66(1): 53-60.
DOI:10.1093/mollus/66.1.53 |
Van Oosterhout C, Hutchinson W F, Wills D P M, Shipley P. 2004. MICRO-CHECKER:software for identifying and correcting genotyping errors in microsatellite data. Mol.Ecol. Notes, 4(3): 535-538.
DOI:10.1111/j.1471-8286.2004.00684.x |
Zeng T, Yin W, Xia R, Fu C Z, Jin B S. 2015. Complete mitochondrial genome of a freshwater snail, Semisulcospira libertina (Cerithioidea:Semisulcospiridae). Mitochondr.DNA, 26(6): 897-898.
DOI:10.3109/19401736.2013.861449 |
 2020, Vol. 38
2020, Vol. 38