Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CEN Jingyi, WANG Jianyan, HUANG Lifen, DING Guangmao, QI Yuzao, CAO Rongbo, CUI Lei, LÜ Songhui
- Who is the "murderer" of the bloom in coastal waters of Fujian, China, in 2019?
- Journal of Oceanology and Limnology, 38(3): 722-732
- http://dx.doi.org/10.1007/s00343-019-9178-6
Article History
- Received Jul. 8, 2019
- accepted in principle Jul. 30, 2019
- accepted for publication Sep. 8, 2019
2 Key Laboratory of Aquatic Eutrophication and Control of Harmful Algae Blooms of Guangdong Higher Education Institutes, Guangzhou 510632, China;
3 Department of Science Research, Beijing Museum of Natural History, Beijing 100050, China;
4 Fujian Fishery Resources Monitoring Center, Fuzhou 350003, China
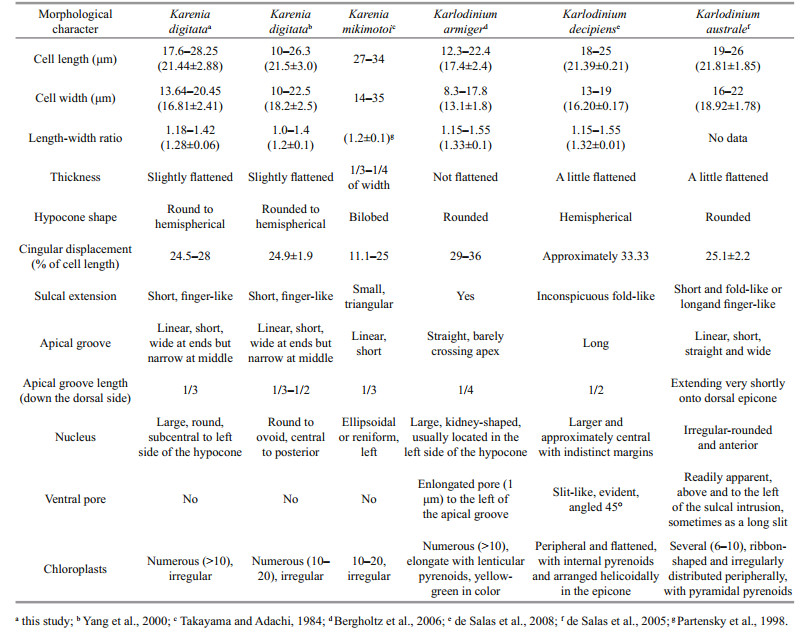
Harmful algal blooms are proliferations of algae that kill marine organisms, contaminate seafood with toxins, and/or cause ecological damage (GEOHAB, 2001). Karenia G. Hansen & Moestrup 2000, a harmful genus of unarmored dinoflagellates, was established following a molecular and morphological study of athecate dinoflagellates previously contained within the genus Gymnodinium (Daugbjerg et al., 2000; Gómez et al., 2005). Major characteristics of Karenia are the presence of a straight apical groove on the epicone and the possession of fucoxanthin or fucoxanthin-derived accessory pigments (de Salas et al., 2003; Bergholtz et al., 2006).
In the past few decades, Karenia blooms have occurred in many sea areas worldwide, such as the United States (Tester and Steidinger, 1997), New Zealand (Chang et al., 2001), Japan (Oda, 1935), Korea (Cho, 1981), China (Lu and Hodgkiss, 2004; Lu et al., 2014), Norway (Tangen, 1977), Chile (Clément et al., 2001), Australia (de Salas et al., 2004), and South Africa (Botes et al., 2003). Karenia species have become well known because the toxins they produced can cause animal mass mortalities, neurotoxic shellfish poisoning, and respiratory distress (Gentien, 1998; Heil and Steidinger, 2009; Brand et al., 2012).
Blooms of Karenia species are also severe and frequent occurred in China. K. mikimotoi is widely distributed in Chinese coastal waters, and was the second-most devastating species in this country (Lu and Hodgkiss, 2004; Lu et al., 2014). According to the China Marine Disaster Bulletin, 2005–2017, the economic loss induced by K. mikimotoi blooms was approximately US$377 million in China (SOAC, 2005–2017). Notably, in 2012 alone, 12 consecutive blooms of K. mikimotoi occurred in the coastal waters of Zhejiang and Fujian provinces, and caused massive financial losses exceeding US$330 million (SOAC, 2013; Li et al., 2017).
Currently, 12 described species have been identified in the genus Karenia (Gómez, 2012), and only 4 species (K. mikimotoi, K. brevis, K. digitata, and K. longicanalis) have been reported in Chinese coastal waters (Liu, 2008). Although only K. mikimotoi was highly concerned and clearly studied for its frequent bloom events in China, we have detected other species, eg. K. longicanalis, was co-occurred with K. mikimotoi in some blooms (Wang et al., 2018). Multi-species Karenia blooms were also observed in other foreign coastal waters (Haywood et al., 2004; Steidinger et al., 2008; Heil and Steidinger, 2009). Different Karenia species have different toxin profiles and contribute differently to fish or shellfish mortality (Brand et al., 2012). Therefore, it is necessary to perform a comprehensive and accurate identification for the causative species, especially in a harmful and toxic algal bloom, and this is also useful for the bloom management and monitoring.
During May 24–29, 2019, a bloom occurred in Pingtan coastal areas of Fujian province. Two species were mainly occurred in the bloom, the first dominated species was identified as Prorocentrum donghaiense, with a concentration attached 1.46×107 cells/L (Cen, unpublished), and the second-most dominant species, with a concentration as high as 4.58×106 cells/L, was another unknown naked dinoflagellate. The bloom caused massive mortality of cage-cultured fish (Plectorhinchus cinctus and Pagrosomus major) (reported by Fujian Provincial Department of Ocean and Fisheries, 2019). In this study, we will give a detailed identification of the unknown dinoflagellate based on morphological observations and a molecular sequence analysis, aiming to explore the "murderer" who killed the massive culture-fish.
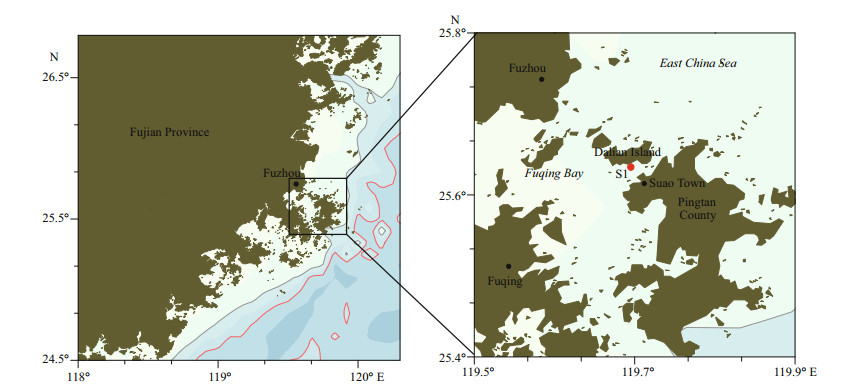
2 MATERIAL AND METHOD 2.1 Phytoplankton samplingSeawater samples were collected on May 26, 2019, during the bloom in Pingtan coastal area, Fujian Province (26°22'39''N, 119°52'42''E) (Fig. 1, station S1). Two strains of the dominated unknown naked dinoflagellate (PT-A and PT-B) were established by isolating single cell from the bloomed seawater samples. The cultures were maintained at 20℃ in L1 medium (Guillard and Hargraves, 1993) at a salinity of 30, under 50 μmol photons/(m2·s) irradiance and a 12-h:12-h light:dark cycle. The cultures were deposited in the Research Center for Harmful Algae and Marine Biology, Jinan University, Guangzhou, China.
|
| Fig.1 Map of Fujian Province (left) showing sampling sites (right) |
Both live field samples and lab cultures were observed under Olympus BX61 microscope (Olympus, Tokyo, Japan). Photographs were captured using the microscope equipped with a QImaging Retiga 4000R digital camera (QImaging, Surrey, BC, Canada). Cell nuclei and chloroplasts were visualized by epifluorescence microscopy. For epifluorescence micrographs, 1-mL portions of field samples were fixed with SYBR Safe DNA (Thermo Fisher, Waltham, MA, USA), and photographed with 360– 370 nm excitation and 420–460 nm emission filters.
2.3 Cell measurementsLengths, widths, and the degree of girdle displacement of 88 randomly selected cultured cells were measured using Image-Pro Plus 6.0 image acquisition and analysis software (QImaging) on a PC coupled to an Olympus BX61 inverted light microscope.
2.4 Scanning electron microscopy (SEM)For SEM observations, 5-mL of each bloom sample was fixed in a final concentration of 2.5% glutaraldehyde for 12 h at 4℃. The fixed culture cells were then filtered onto a Whatman filter (8 μm) and rinsed in deionized water for 10 min. Samples were dehydrated using a 10%, 30%, 50%, 70%, and 90% ethanol series, 20 min at each change, followed by three changes for 10 min each in 100% ethanol. The samples were subsequently critical-point dried in liquid CO2 using a Leica EM CPD300 critical point dryer (Leica Microsystems, Mannheim, Germany). After gold-palladium coating, mounted samples were observed under an Ultra 55 field emission scanning electron microscope (Zeiss, Jena, Germany).
2.5 PCR amplification of LSU and ITS DNA regionsFor field samples, single cell was isolated from the seawater and transferred into 200-μL PCR tubes for PCR amplification. For lab cultures, 2-mL of the cell culture was collected by centrifugation, and total genomic DNA was extracted using a TaKaRa MiniBEST Universal Genomic DNA Extraction Kit (TaKaRa, Dalian, China) according to the manufacturer's protocol. The D1–D3 region of the LSU of rDNA (~850 base pairs) was amplified using primers D1R (Scholin et al., 1994) and D3Ca (Lenaers et al., 1989), and the ITS region was amplified with primers ITS1 and ITS4 (Stern et al., 2012). PCR amplifications were carried out in a PTC-200 Peltier thermal cycler (MJ Research, San Francisco, CA, USA). Cycling conditions for amplification of the LSU were 94℃ for 3 min, followed by 35 cycles of 94℃ for 1 min, 55℃ for 1 min 30 s, and 72℃ for 1 min, with a final extension at 72℃ for 10 min. The protocol for ITS amplification was as follows: 94℃ for 3 min, followed by 34 cycles of 94℃ for 30 s, 47℃ for 30 s, and 72℃ for 45 s, with a final extension at 72℃ for 7 min. The PCR products were purified and sent to Beijing Genomics Institute (Guangzhou, China) for sequencing.
2.6 Sequence alignment and phylogenetic analysesThe generated sequences were aligned in ClustalX (Thompson et al., 1997) with other representative Kareniaceae downloaded from GenBank, followed by manual refinements. The general accepted maximum likelihood and Bayesian inference methods were carried out to analyze the phylogenetic relationship among the relevant species, using Gymnodinium catenatum and Heterosigma akashiwo as outgroup. The maximum likelihood analysis was carried out in MEGA version 6.0 (Tamura et al., 2013), and Bayesian inference was performed in MrBayes 3.1.2 after determination of the best-fitting model (Ronquist and Huelsenbeck, 2003). Statistical support for branches in the ML and BI trees was assessed using 1 000 bootstrap iterations.
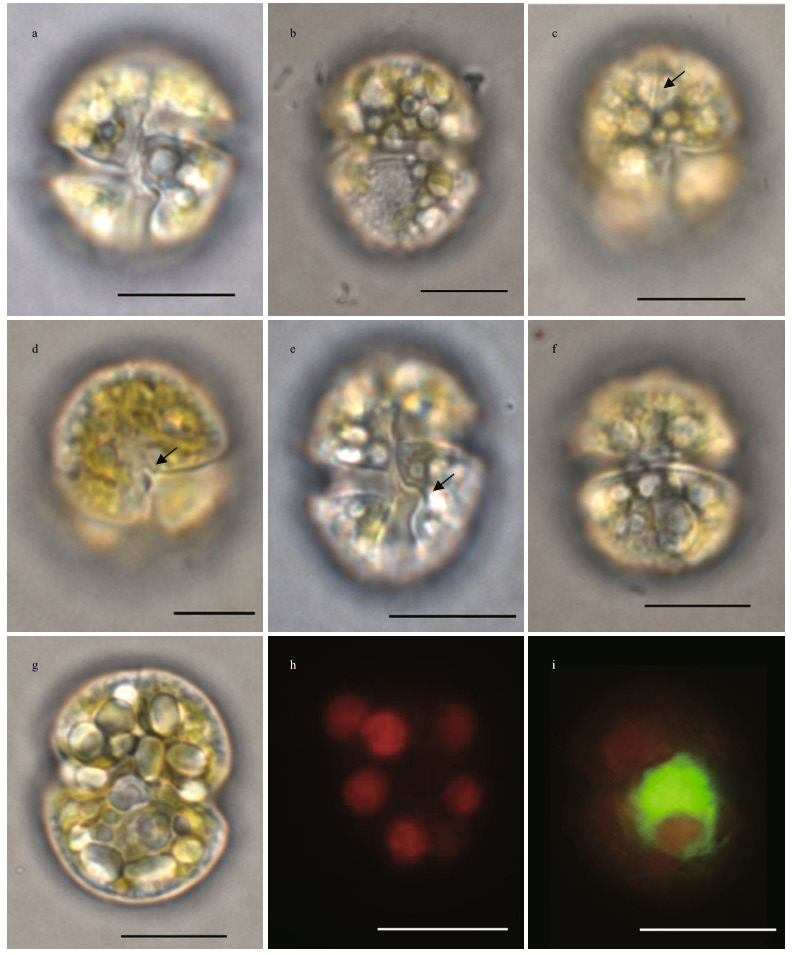
3 RESULT 3.1 Morphology and characterization of the unknown causative dinoflagellate 3.1.1 LMCells appeared globular or oval in shape (Fig. 2a & b), with a mean length of 20.34±1.54 μm (n=88), a mean width of 16.02±1.3 μm (n=88), and a length:width ratio of 1.27±0.06 (n=88). The straight apical groove extended from the dorsal apex to the ventral epicone, and located at the right side of the sulcal axis (Fig. 2c). In the ventral view, the sulcus invaded the epicone slightly as a small finger-like extension (Fig. 2d). When meeting the cingulum, the sulcus turned to left and formed curved knot on the left lobe of the hypocone (Fig. 2e). The cingulum was relatively wide, median, and encircled the cell equatorially (Fig. 2f).The chloroplasts were yellowgreen to yellowish brown, spherical in shape, and irregularly distributed in cells (Fig. 2g & h). The large spherical nucleus was located centrally to posteriorly (Fig. 2i). Cells moved smoothly and quickly in a rotating manner.
|
| Fig.2 Light and epifluorescence micrographs of Karenia digitata a. cell in ventral view; b. cell in dorsal view; c. ventral view, showing the straight apical groove (arrow); d. ventral view, showing the sulcus invaded the epicone (arrow); e. ventral view, showing the structure of the curve knot (arrow); f. dorsal view, showing the cingulum; g. ventral view, showing the chloroplasts of yellow-green to yellowish-brown; h. epifluorescence micrograph showing the peripheral, spherical chloroplasts; i. SYBR-stained cell showing the position of the nucleus (green). Scale bar=10 μm. |
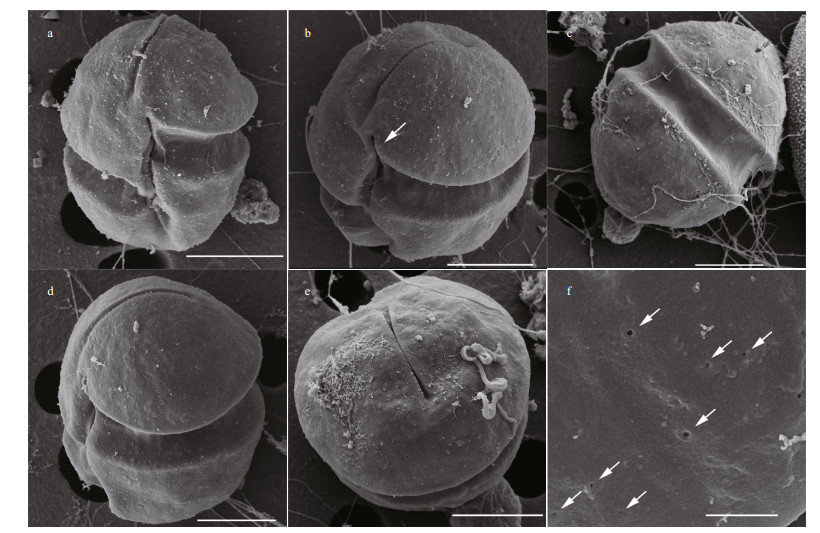
Observed under SEM, the epicone and hypocone were both hemispherical and were approximately equally sized (Fig. 3a). The sulcus extended from just above the proximal end of the cingulum and elongated nearly to the antapex end. The linear apical groove was distinct under SEM, and located at the right of the sulcal axis. The cingulum displacement was approximately 25% of the total cell length (Fig. 3a). The sulcus intruded onto the epicone as a finger-like projection (Fig. 3a & b). The hypocone was conical, and did not notch by the sulcus at the dorsal side (Fig. 3c). The apical groove was narrow and straight, extending about one-third of the length to the dorsal side of the epicone (Fig. 3c & d). The apical groove was narrow in center, and became wide at the two ends (Fig. 3e). Cell surfaces had irregularly distributed pores, mainly on the epicone (Fig. 3f).
|
| Fig.3 Scanning electron micrographs of Karenia digitata a. ventral views, showing the cells as globular in shape; b. ventral oblique views, showing the finger-like sulcal intrusion (arrow); c. dorsal view, showing the apical groove extension and the dorsal side of hypocone; d. lateral oblique views, showing the linear apical groove extending from the ventral to the dorsal side of eipcone; e. top view of cell, showing the apical groove; f. details of the irregularly distributed pores on the epicone (arrowhead). a–e: scale bar=5 μm; f: scale bar=1 μm. |
Based on the observation under LM and SEM, and the comparison with other morphologically similar species (Table 1), the unknown naked dinoflagellate was featured by a straight apical groove on the epicone, an obvious finger-like extension of the sulcus into the epicone, and a sulcal curvature on the left lobe of hypcone. The morphological characteristics of the unknown dinoflagellate were exactly in accordance with the typical features of K. digitata described in Yang et al. (2000); therefore, we identified the unknown causative species as K. digitata.
LSU sequences of four single-cell PCR products (PT01, PT02, PT03, and PT04) and two culture strains of the causative species K. digitata (PT-A and PT-B) were obtained and deposited in GenBank under accession numbers MN134472 to MN134476, and MN134478. No molecular sequence of K. digitate was provided by Yang et al. (2000) when this species was first reported in 2000. In 2011, Lee et al. (2011) released the LSU and ITS rDNA sequences of a species isolated in Hong Kong, China, which they identified as K. digitata (stain No. KD01). The LSU sequences of Fujian cultures in the present study were nearly identical to that of K. digitata reported in Lee et al. (2011), with only one difference observed among the total 704 base pairs.
ITS sequences of four single-cell PCR products (PT-05, PT-06, PT-07, and PT-08) and two culture strains of the causative species K. digitata (PT-A and PT-B) were obtained and deposited in GenBank under accession numbers MN133930 to MN133934, and MN309841. The ITS sequences of the four single-cell PCR products (615 base pairs) were identical to that of the KD01 strain of K. digitata reported by Lee et al. (2011), and the two cultures (PT-A and PT-B) differed from the strain KD01 by 2 bases. Therefore, the LSU rDNA and ITS rDNA sequences also proved the fact that the causative dinoflagellate was K. digitata.
3.2.2 Phylogenetic analysisIn the phylogenetic tree based on LSU rDNA sequences, K. digitata isolated in the present study were clustered with K. digitata KD01, and formed a single clade with bootstrap/posterior probability values of 96%/0.95 (Fig. 4). The clade of K. digitata was most closely related to Karl. austrael and Karl. armiger, and the three species clustered into a single branch with bootstrap/posterior probability values of 95%/0.99 (Fig. 4). The phylogenetic tree constructed on ITS rDNA sequences outputted the similar results as LSU rDNA tree, and the Pingtan strains of K. digitata and Hong Kong strain (KD01) were conspecific, closely clustering to Karl. australe and Karl. armiger (Fig. 5).
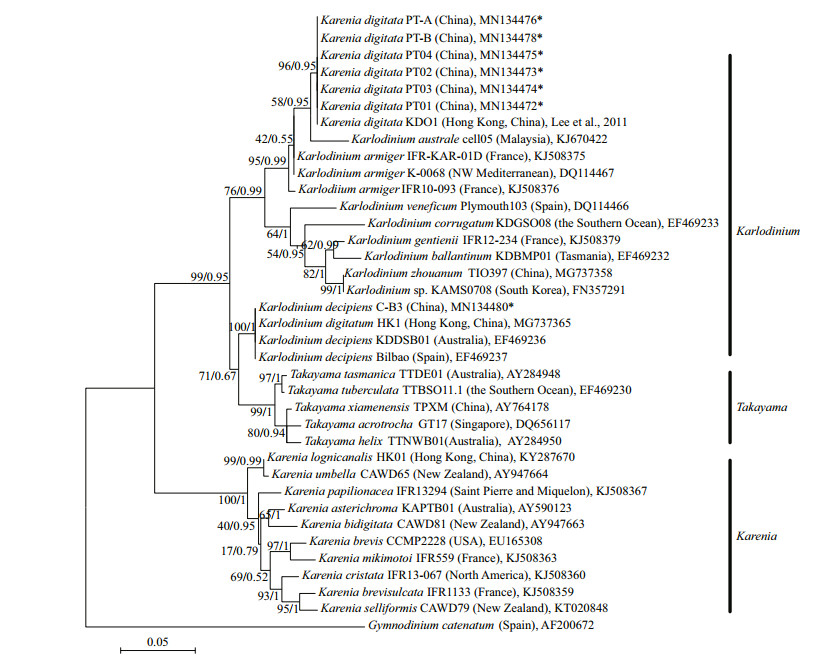
|
| Fig.4 Maximum-likelihood phylogenetic tree of LSU sequences of Karenia digitata and closely related species Gymnodinium catenatum was used as an out-group. GenBank accession numbers and geographical origins are given for each sequence. Sequences generated in this study were marked with asterisk. Values at branches were maximum-likelihood bootstrap percentages and Bayesian posterior probabilities. Only values ≥50/0.80 were shown. |
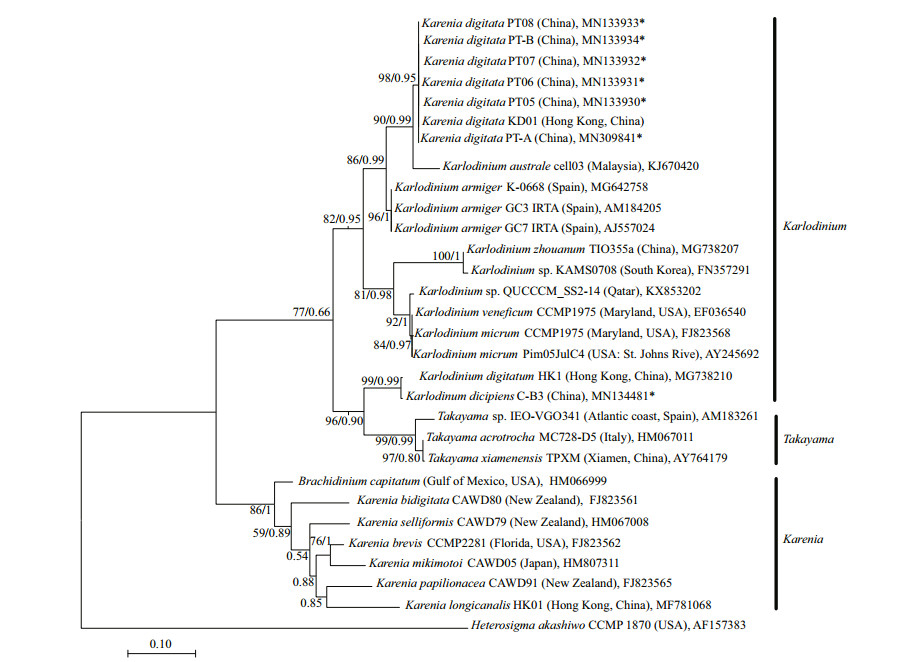
|
| Fig.5 Maximum-likelihood phylogenetic tree of ITS sequences of Karenia digitata and closely related species Heterosigma akashiwo was used as an out-group. GenBank accession numbers and geographical origins are given for each sequence. Sequences generated in this study were marked with asterisk. Values at nodes were maximum likelihood bootstrap values and Bayesian posterior probabilities. Only values ≥50/0.80 were shown. |
In the tree inferred from LSU rDNA, the 10 Karenia species (K. longicanalis, K. umbella, K. papilionacea, K. asterichroma, K. bidigitata, K. brevis, K. mikimotoi, K. cristata, K. brevisulcata, and K. selliformis) formed a clearly single branch (with bootstrap/posterior probability values of 100%/1), while the Pingtan K. digitata as well as Hong Kong K. digitata (KD01) was strongly clustered with Karlodinium species (Karl. australe, Karl. armiger, Karl. zhouanum, Karl. veneficum, and Karl. micrum) with bootstrap/posterior probability values of 76%/0.99. The ITS tree also revealed the similar results with LSU rDNA that species belonging to Karenia genus formed a clade (86%/1), while K. digitata and some Karlodinium species clustered into another clade (82%/0.95). The results suggested that K. digitata was phylogenetically belonged to the genus Karlodinium rather than genus Karenia.
3.3 Revison of Karenia digitata as Karlodinium digitatumBased on the morphological characteristics, the naked causative species was identified as K. digitata. According to the phylogenetic data, this species was classified to genus Karlodinium. Luo et al. (2018) performed a re-taxonomy of "K. digitata" (strain HK1) and emended it to Karlodinium digitatum based on the LSU rDNA and ITS sequences. Although maybe based on a culture of Karl. decipiens, the combination of Karl. digitatum Gu, Chan & Lu in Luo et al. (2018) is valid and this is the correct name of the original organism K. digitata. Therefore, the naked causative dinoflagellate in Pingtan coastal areas of Fujian Province, on May 24–29, 2019, was finally identified as Karl. digitatum.
4 DISCUSSIONKarenia digitata was first described and placed in the genus Karenia because of its straight apical groove and the finger-like extension of the sulcus into the epicone (Yang et al., 2000). However, K. digitata more closely resembles Karlodinium species than those of Karenia in regards to body size and other morphological features (Table 1). The absence of a ventral pore was originally used to distinguish K. digitata from members of the genus Karlodinium, although species lacking a ventral pore, such as Karl. ballantinum (de Salas et al., 2008) and Karl. zhouanum (Luo et al., 2018), have subsequently been found to belong in Karlodinium. The nucleus of K. digitata is large and round, and is sub-centrally located on the posterior and left side of the hypocone; this nuclear location is different from that of Karl. ballantinum (centrally located on the dorsal cell surface) and Karl. zhouanum (located in the lower epicone).
The sulcus extension into the epicone has also been observed in other Karenia and Karlodinium species. In most Karenia species, the sulcus extension invades the epicone as a small closed or open-ended triangular extension (Yang et al., 2000, Figures 18–27; Haywood et al., 2004, Figure 6), and only one species, K. umbella, has a finger-like sulcus extension like that of K. digitata (de Salas et al., 2004). Karenia umbella can be distinguished from K. digitata by its large cell size and the six or eight radial furrows on the epicone that are visible by SEM (Wang et al. (2018) concluded that K. umbella and K. longicanalis are the same species). Sulci in most species of Karlodinium extend into the epicone, and some, such as Karl. armiger, Karl. australe, Karl. conicum, Karl. corrugatum, and Karl. gentienii have a finger-like shape; however, the species with a finger-like sulcus intrusion all have an apparent ventral pore that allows them to be easily discriminated from K. digitata.
When K. digitata was first reported, DNA sequences of the holotype were not available. Lee et al. (2011) published the LSU and ITS sequence of culture cells of K. digitata isolated from algal bloom samples collected in Silver Mine Bay during March 23–26, 2009 (The identification of the causative agent as K. digitata was confirmed morphologically by officials of the Agricultural, Fisheries and Conservation Department of the Hong Kong SAR Government). In 2018, Luo et al. (2018) published LSU and ITS sequences of bloom samples they collected and freeze dried during the bloom event in 1998, which they identified as "K. digitata" and emended to Karl. digitatum. The ML and BI phylogenetic analyses of LSU and ITS sequences in the present study suggested that the Pingtan strains of K. digitata are conspecific with the Hong Kong KD01 strain of Lee et al. (2010), and they clustered with species of Karlodinium in the phylogenetic trees; while the "K. digitata" (HK1) reported by Luo et al. (2018) was very similar to Karlodinium decipiens, clustering with the latter in the LSU-based tree with bootstrap/posterior probability values of 100%/1.0, and formed a subclade with Takayama. In their discussion, Luo et al. (2018) suggested that LSU sequences may be too conserved to differentiate this group of Kareniaceae species, and this is the reason why "K. digitata" and Karl. decipiens have identical LSU sequences even though they are significantly distinct in morphology. Because there was no morphological description of "K. digitata" (HK1) when they was pressed by Luo et al. (2018), we proposed that the sequences cannot be confidently assigned to the bloom-causative organism K. digitata in 1998, but more likely is a strain of Karl. decipiens. The molecular sequences representing Karl. digitatum, however, are those of Lee et al. (2011) and those published in the present study.
In the present study, the causative naked species in Pingtan bloom was emended as Karl. digitatum from the original K. digitata. The genera Karenia and Karlodinium both have a straight apical groove on epicone. Morphologically, Karenia species are more dorsoventrally flattened than Karlodinium species (Haywood et al., 2004). A key characteristics distinguishing Karlodinium from Karenia is the presence of a ventral pore in Karlodinium (Bergholtz et al., 2005, de Salas et al., 2008). However, the increasing discoveries of species, which lack a ventral pore but are genetically consistent with Karlodinium, e.g. Karl. zhouanum, and Karl. digitatum in the present study, raise the issue of what remains to differentiate Karlodinium from Karenia.
Karenia mikimotoi was the most common harmful bloom species in Fujian coastal areas, China (Xu et al., 2010). K. mikimotoi is much bigger and more flattened than Karl. digitatum in body appearance. The hypocone of K. mikimotoi was sliced to bilobeshaped by the sulcus, while that of Karl. digitatum was rounded to semispherical (Yang et al., 2000; Haywood et al., 2004). The naked cell of Karenia species is easy to be deformed after fixation when the cells are checked under LM, and this may lead to the misidentification of causative species. It is uncertain whether the actual causative species of "Karenia" was K. mikimotoi in the past years in Fujian coast.
In 1998, the Karl. digitatum (basionym: K. digitata) bloom invaded 22 of Hong Kong's 26 coastal fish farms, killed 90% of caged fish, and caused losses of approximately HK$250 million (US$32 million) in Hong Kong, China (Yang and Hodgkiss, 1999, 2004). This species apparently first appeared in Hong Kong in 1989 and then again in 1995, but it did not form a bloom until 1998. Karl. digitatum is suspected to have bloomed along the coasts of western Japan in the summers of 1995 and 1996 and in November 1997. Various fish species, such as red sea bream (Pagrus major), bastard halibut (Paralichthys olivaceus), sea bass (Lateolabrax japonicus), file fish (Acanthurus xanthopterus), yellow tail (Seriola quinqueradiata), and horse mackerel (Trachurus japonicus), were reportedly killed by this naked dinoflagellate (Landsberg, 2002). Moreover, cultured seaweeds (Porphyra tenera) were affected by the bloom in Japan and exhibited irregular cell growth, which suggests that this alga is also harmful to marine plants. Strangely and interestingly, no further studies of this species were conducted until 2009 by Lee et al. (2011), and very few data have been collected worldwide on this harmful species until now.
During the bloom in coastal waters of Fujian, China, in 2019, P. donghaiense and Karl. digitatum were the most abundant species. P. donghaiense has formed high biomass in the East China Sea since 1998; however, it is neither a toxin producer nor a species associated with fish kills (Lu et al., 2014). As mentioned above, huge mortality of fish has been caused by Karl. digitatum in Hong Kong and Japan, we supposed that the "murderer" who killed the massive culture-fish in the bloom during May 24–29, 2019 in Pingtan coastal areas of Fujian province was Karl. digitatum.
A notable feature of Karl. digitatum collected from Pingtan is the many irregularly placed pores on the cell surface that are visible under SEM (Fig. 3d). This feature was not mentioned in Yang et al. (2000), however, de Salas et al. (2005) found a row of small processes on the hypocone of K. digitata described in Yang et al. (2000) (Figures 10–12). Similar structure, trichocysts pores were observed in Karl. armiger (Bergholtz et al., 2006), a toxic species which can producd karmitoxin and kill copepod (Rasmussen et al., 2017). We hypothesis that the pores distributed on the cell surface of Karl. digitatum are those of trichocysts and are related to toxin production.
5 CONCLUSIONIn this study, the second causative organism of the bloom in Fujian coastal waters, China, in 2019 was isolated and identified. On the basis of its morphological characteristics and molecular sequences, the species was identified as Karl. digitatum and found to be conspecific with K. digitata which bloomed in the coastal waters of western Japan and Hong Kong in 1998. We proposed that the massive mortality of fish in this bloom was induced by the toxic species Karl. digitatum.
6 DATA AVAILABILITY STATEMENTThe data that support the findings of this study are available from the corresponding author upon request.
7 ACKNOWLEDGMENTSpecial appreciation is expressed to Prof. Moestrup of the University of Copenhagen for helpful discussions regarding the species identification. We also sincerely thank Dr. ZHEN Yu of the Ocean University of China for his assistance with the molecular data analysis.
Bergholtz T, Daugbjerg N, Moestrup Ø, Fernández-Tejedor M. 2006. On the identity of Karlodinium veneficum and description of Karlodinium armiger sp. nov.(Dinophyceae), based on light and electron microscopy, nuclear-encoded LSU rDNA, and pigment composition. Journal of Phycology, 42(1): 170-193.
|
Botes L, Smit A J, Cook P A. 2003. The potential threat of algal blooms to the abalone (Haliotis midae) mariculture industry situated around the South African coast. Harmful Algae, 2(4): 247-259.
|
Brand L E, Campbell L, Bresnan E. 2012. Karenia:the biology and ecology of a toxic genus. Harmful Algae, 14(1): 156-178.
|
Chang F H, Chiswell S M, Uddstrom M J. 2001. Occurrence and distribution of Karenia brevisulcata (Dinophyceae)during the 1998 summer toxic outbreaks on the central east coast of New Zealand. Phycologia, 40(3): 215-221.
|
Cho C H. 1981. On the Gymnodinium red tide in Jinhae Bay. Korean Journal of Fisheries and Aquatic Sciences, 14(4): 227-232.
|
Clément A, Seguel M, Arzul G, Guzmán L, Alarcón C. 2001. A widespread outbreak of a haemolytic, ichthyotoxic Gymnodinium sp. in southern Chile. In: Hallegraeff G M, Blackburn S I, Bolch C J, Lewis R J eds. Harmful algal blooms 2000. Intergovernmental Oceanographic Commission, UNESCO, Paris. p.66-69.
|
Daugbjerg N, Hansen G, Larsen J, Moestrup Ø. 2000. Phylogeny of some of the major genera of dinoflagellates based on ultrastructure and partial LSU rDNA sequence data, including the erection of three new genera of unarmoured dinoflagellates. Phycologia, 39(4): 302-317.
|
de Salas M F, Bolch C J S, Botes L, Nash G, Wright S W, Hallegraeff G M. 2003. Takayama gen. nov.(Gymnodiniales, Dinophyceae), a new genus of unarmored dinoflagellates with sigmoid apical grooves, including the description of two new species. Journal of Phycology, 39(6): 1 233-1 246.
|
de Salas M F, Bolch C J S, Hallegraeff G M. 2004. Karenia umbella sp. nov. (Gymnodiniales, Dinophyceae), a new potentially ichthyotoxic dinoflagellate species from Tasmania, Australia. Phycologia, 43(2): 166-175.
|
de Salas M F, Bolch C J S, Hallegraeff G M. 2005. Karlodinium australe sp. nov. (Gymnodiniales, Dinophyceae), a new potentially ichthyotoxic unarmoured dinoflagellate from lagoonal habitats of south-eastern Australia. Phycologia, 44(6): 640-650.
|
de Salas M F, Laza-Martínez A, Hallegraeff G M. 2008. Novel unarmored dinoflagellates from the toxigenic family Kareniaceae (Gymnodiniales):five new species of Karlodinium and one new Takayama from the Australian sector of the Southern Ocean. Journal of Phycology, 44(1): 241-257.
|
Fujian Provincial Department of Ocean and Fisheries, China. 2019. Red tide disaster information in Fujian Province(No. 019). http://hyyyj.fujian.gov.cn/xxgk/tzgg/201905/t20190525_4885308.htm
|
Gentien P. 1998. Bloom dynamics and ecophysiology of the Gymnodinium mikimotoi species complex. In: Anderson D M, Cembella A D, Hallegraeff G M eds. Physiological Ecology of Harmful Algal Blooms. Springer, Berlin.p.155-173.
|
GEOHAB. 2001. Global Ecology and Oceanography of Harmful Algal Blooms. In: Gilbert P M, Pitcher G eds. Science Plan.SCOR- IOC (UNESCO), Baltimore and Paris. 86p.
|
Gómez F, Nagahama J, Takayama H, Furuya K. 2005. Is Karenia a synonym of Asterodinium-Brachidinium (Gymnodiniales, Dinophyceae)?. Acta Botanica Croatica, 64(2): 263-274.
|
Gómez F. 2012. A checklist and classification of living dinoflagellates (Dinoflagellata, Alveolata). Cicimar Oceánides, 27(1): 65-140.
|
Guillard R R L, Hargraves P E. 1993. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia, 32(3): 234-236.
|
Haywood A J, Steidinger K A, Truby E W, Bergquist P R, Bergquist P L, Adamson J, Mackenzie L. 2004. Comparative morphology and molecular phylogenetic analysis of three new species of the genus Karenia(Dinophyceae) from New Zealand. Journal of Phycology, 40(1): 165-179.
|
Heil C A, Steidinger K A. 2009. Monitoring, management, and mitigation of Karenia blooms in the eastern Gulf of Mexico. Harmful Algae, 8(4): 611-617.
|
Landsberg J H. 2002. The effects of harmful algal blooms on aquatic organisms. Reviews in Fisheries Science, 10(2): 113-390.
|
Lee F W, Ho K C, Mak Y L, Lo C L. 2011. Authentication of the proteins expression profiles (PEPs) identification methodology in a bloom of Karenia digitata, the most damaging harmful algal bloom causative agent in the history of Hong Kong. Harmful Algae, 12: 1-10.
|
Lenaers G, Maroteaux L, Michot B, Herzog M. 1989. Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. Journal of Molecular Evolution, 29(1): 40-51.
|
Li X D, Yan T, Lin J N, Yu R C, Zhou M J. 2017. Detrimental impacts of the dinoflagellate Karenia mikimotoi in Fujian coastal waters on typical marine organisms. Harmful Algae, 61: 1-12.
|
Liu R Y. 2008. Checklist of Marine Biota of China Seas. Science Press, Beijing, China. p.177.
(in Chinese)
|
Lu D D, Qi Y Z, Gu H F, Dai X F, Wang H X, Gao Y H, Shen P P, Zhang Q C, Yu R C, Lu S H. 2014. Causative species of harmful algal blooms in Chinese coastal waters. Algological Studies, 145-146(1): 145-168.
|
Lu S H, Hodgkiss I J. 2004. Harmful algal bloom causative collected from Hong Kong waters. Hydrobiologia, 512(1-3): 231-238.
|
Luo Z H, Wang L, Chan L, Lu S H, Gu H H. 2018. Karlodinium zhouanum, a new dinoflagellate species from China, and molecular phylogeny of Karenia digitata and Karenia longicanalis (Gymnodiniales, Dinophyceae). Phycologia, 57(4): 401-412.
|
Oda M. 1935. Gymnodinium mikimotoi Miyake et Kominami n. sp. (MS) and the influence of copper sulfate on the red tide. Dobutsugaku Zassh, 47: 35-48.
|
Partensky F, Vaulot D, Couté A, Sournia A. 1988. Morphological and nuclear analysis of the bloom-forming dinoflagellates Gyrodinium cf.aureolum and Gymnodinium nagasakiense. Journal of Phycology, 24(3): 408-415.
|
Rasmussen S A, Binzer S B, Hoeck C, Meier S, de Medeiros L S, Andersen N G, Place A, Nielsen K F, Hansen P J, Larsen T O. 2017. Karmitoxin:an amine-containing polyhydroxy-polyene toxin from the marine dinoflagellate Karlodinium armiger. Journal of Natural Products, 80(5): 1 287-1 293.
|
Ronquist F, Huelsenbeck J P. 2003. MrBayes 3:bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1 572-1 574.
|
Scholin C A, Herzog M, Sogin M, Anderson D M. 1994. Identification of group-and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). Ⅱ.Sequence analysis of a fragment of the LSU rRNA gene. Journal of Phycology, 30(6): 999-1 011.
|
SOAC 2005-2017 (State Oceanic Administration, China). The National Marine Economic Statistics Bulletin, China Oceanic Information Network (2005-2017). http://m.lc.mlr.gov.cn/sj/sjfw/hy/gbgg/zghyzhgb/.(in Chinese)
|
Steidinger K A, Wolny J L, Haywood A J. 2008. Identification of kareniaceae (Dinophyceae) in the gulf of Mexico. Nova Hedwigia, 133: 269-284.
|
Stern R F, Andersen R A, Jameson I, Küpper F C, Coffroth M A, Vaulot D, Le Gall F, Véron B, Brand J J, Skelton H, Kasai F, Lilly E L, Keeling P J. 2012. Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS One, 7(8): e42780.
|
Takayama H, Adachi R. 1984. Gymnodinium nagasakiense sp.nov., a red-tide forming dinophyte in the adjacent waters of Japan. Bulletin of Plankton Society of Japan, 31(1): 7-14.
|
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2 725-2 729.
|
Tangen K. 1977. Blooms of Gyrodinium aureolum(Dinophygeae) in north European waters, accompanied by mortality in marine organisms. Sarsia, 63(2): 123-133.
|
Tester P A, Steidinger K A. 1997. Gymnodinium breve red tide blooms:Initiation, transport, and consequences of surface circulation. Limnology and Oceanography, 42(5): 1 039-1 051.
|
Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The Clustal_X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24): 4 876-4 882.
|
Wang J Y, Cen J Y, Li S, Lü S H, Moestrup Ø, Chan K K, Jiang T, Lei X D. 2018. A re-investigation of the bloom-forming unarmored dinoflagellate Karenia longicanalis (syn.Karenia umbella) from Chinese coastal waters. Journal of Oceanology and Limnology, 36(6): 2 202-2 215.
|
Xu C Y, Huang M Z, Du Q. 2010. Ecological characteristics of important red tide species in Fujian coastal waters. Journal of Oceanography in Taiwan Strait, 29(3): 434-441.
(in Chinese with English abstract) |
Yang Z B, Hodgkiss I J. 1999. Massive fish killing by Gyrodinium sp. Harmful Algae News, 18: 4-5.
|
Yang Z B, Hodgkiss I J. 2004. Hong Kong's worst "red tide"-causative factors reflected in a phytoplankton study at Port Shelter station in 1998. Harmful Algae, 3(2): 149-161.
|
Yang Z B, Takayama H, Matsuoka K, Hodgkiss I J. 2000. Karenia digitata sp. nov. (Gymnodiniales, Dinophyceae), a new harmful algal bloom species from the coastal waters of west Japan and Hong Kong. Phycologia, 39(6): 463-470.
|
 2020, Vol. 38
2020, Vol. 38