Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LIU Changshui, ZANG Kun, LI Shihao, LI Fuhua, MA Qingjun
- Structural analysis of a shrimp thymidylate synthase reveals species-specific interactions with dUMP and raltitrexed
- Journal of Oceanology and Limnology, 38(6): 1891-1899
- http://dx.doi.org/10.1007/s00343-019-9184-8
Article History
- Received Jul. 16, 2019
- accepted in principle Aug. 29, 2019
- accepted for publication Nov. 12, 2020
2 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266000, China;
3 University of Chinese Academy of Sciences, Beijing 100049, China;
4 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
Thymidylate synthase (TS) is one of the most evolutionarily conserved proteins in all organisms (Perry et al., 1990). TS catalyzes the reductive methylation of deoxyuridine monophosphate (dUMP) to produce thymidine monophosphate (TMP), using 5, 10-methylenetetrahydrofolate as the methyl group donor (Carreras and Santi, 1995). This is an essential step in DNA biosynthesis and inhibiting TS can lead to thymineless death (Jackman, 1999). For a long time, TS has served as a drug target in cancer chemotherapy, with available clinical drugs, such as 5-fluorouracil and raltitrexed (Rustum et al., 1997).
TSs from pathogens have also been proposed as a promising drug target against infectious diseases (de Clercq et al., 1981; Stout et al., 1999). Due to the high similarity between pathogen and host TSs, developing drugs specifically targeting pathogen TSs is challenging. Nevertheless, it is feasible to develop pathogen-specific inhibitors with species selectivity of up to 100-fold (Stout et al., 1999; Zaware et al., 2013). Previous studies showed that human TS (hTS) ternary complex with dUMP and raltitrexed adopts a different conformation from those of TSs of human pathogens such as Mycobacterium tuberculosis (PDB: 4FOX) and Kaposi's sarcoma-associated herpesvirus (Phan et al., 2001; Choi et al., 2016). These species-specific characteristics would be a key to further optimize drug with high species selectivity. Notably, these structural differences could be minor, and thus accurate structural information of both host and pathogen TSs would be crucial for designing pathogen-specific inhibitors.
Shrimp is one of the major aquaculture species. The global annual economic value of shrimp aquaculture has reached more than 30 billion USD (Food and Agriculture Organization of the United Nations, 2018). In recent years, shrimp aquaculture has seriously suffered from diseases caused by viruses and bacteria such as white spot syndrome virus and vibrio (Flegel, 2012; Lightner et al., 2012). For these diseases, either no reliable treatment is available yet, or drug resistance is becoming a big problem. Novel chemotherapy is urgently needed and TS is supposed to be an attractive drug target (Arvizu-Flores et al., 2009). Despite a high structural similarity among TSs, homologous structural models derived from known TS structures failed to reveal structural differences for designing species-specific drugs (Arvizu-Flores et al., 2009). To facilitate effective drug design using TS as a drug target, accurate structural information of both shrimp and pathogen TSs is required.
Here, we report crystal structures of TS from the Pacific white shrimp Litopenaeus vannamei (LvTS), including the apo form, the binary complex with dUMP (LvTS-dUMP), and the ternary complex with dUMP and raltitrexed (LvTS-dUMP-raltitrexed). This is the first report on crustacean TS structure. The structures reveal certain species-specific differences from close homologs, such as hTS and mouse TS (mTS). Particularly, LvTS-dUMP-raltitrexed adopts a loosely closed conformation, with structural features intermediate between the closed and the open conformations reported previously.
2 MATERIAL AND METHOD 2.1 Gene cloning, protein expression and purificationThe DNA sequence encoding LvTS was amplified from a muscle cDNA library of L. vannamei by PCR with primers (forward, 5′-TACTTCCAATCCAATGCCATGAGGCATGACGAGTACCAG-3′; reverse, 5′-TTATCCACTTCCAATGCTATTACACGGCCATTTCCATTTTG-3′), and was inserted into a modified pET30 vector using a ligation independent cloning protocol (Aslanidis and de Jong, 1990). The recombinant protein product was expected to contain an N-terminal 6×His tag followed by a tobacco etch virus protease (TEV) cleavage site. The construct was transformed into Escherichia coli strain Rosetta (DE3). The bacteria were grown in Luria-Bertani broth at 37℃, and then 0.2 mmol/L IPTG was added for induction when OD600 of the culture reached ~0.8, with further culturing at 16℃ for 16 h. Cells were harvested by centrifugation and lysed by sonication in the lysis buffer (50 mmol/L Tris, 150 mmol/L NaCl, 10 mmol/L imidazole, 1 mmol/L DTT, pH 8.0). The lysate was clarified, and the supernatant was loaded onto a Ni-NTA column (GE Healthcare) and eluted in the buffer (50 mmol/L Tris, 150 mmol/L NaCl, 250 mmol/L imidazole, 1 mmol/L DTT, pH 8.0). The elution was changed into the imidazole-free buffer (50 mmol/L Tris, 150 mmol/L NaCl, 1 mmol/L DTT, pH 8.0) using a PD-10 column (GE Healthcare) for later TEV digestion. After the N-terminal 6×His tag was removed, the sample was reloaded onto the NiNTA column to remove uncleaved protein and Histagged TEV. The digested protein was further purified on a Superdex 200 size-exclusion column (HiLoad 16/600, GE Healthcare) in the equilibrium buffer (10 mmol/L HEPES, 150 mmol/L NaCl, 1 mmol/L DTT, pH 7.5). The purified protein was concentrated to about 40 mg/mL determined by absorbance at 280 nm, and was used freshly or frozen at -80℃ for later use. A typical expression and purification profile of the recombinant protein was analyzed on an SDSPAGE gel (Supplementary Fig.S1a), and molecular mass of the recombinant protein was estimated using Superdex 200 Increase 10/300 GL size-exclusion column (GE Healthcare) in the equilibrium buffer (10 mmol/L HEPES, 150 mmol/L NaCl, 1 mmol/L DTT, pH 7.5) (Supplementary Fig.S1b).
2.2 Protein crystallization and structure determinationThe crystallization experiments were carried out at 20℃ using the sitting drop vapor diffusion method. The crystals of apo protein were obtained after 10 days in drops containing 1 μL protein solution (20 mg/ mL) and 1 μL reservoir solution (18% (w/v) PEG 3350, 200 mmol/L (NH4)2SO4, 100 mmol/L HEPES, pH 7.5). The crystals of binary complex with dUMP (Sangon Biotech) grew after 3 days in drops containing 1 μL protein solution (10 mg/mL protein + 5 mmol/L dUMP incubated for 4 h at 4℃) and 1 μL reservoir solution (25% (w/v) PEG 3350, 200 mmol/L Li2SO4, 100 mmol/L Bis-Tris, pH 6.2). The crystals of ternary complex with dUMP and raltitrexed (Wuhu Nuowei Chemical Technology) grew after 3 days in drops containing 1 μL protein solution (10 mg/mL protein + 5 mmol/L dUMP + 5 mmol/L raltitrexed incubated for 4 h at 4℃) and 1 μL reservoir solution (20% (w/v) PEG 3350, 200 mmol/L ammonium citrate, pH 7.0). All the crystals were soaked in corresponding cryobuffers (apo crystal: 35% (w/v) PEG 3350, 200 mmol/L (NH4)2SO4, 100 mmol/L HEPES, pH 7.5; binary complex crystal: 35% (w/v) PEG 3350, 200 mmol/L Li2SO4, 100 mmol/L Bis-Tris, pH 6.2; ternary complex crystal: 35% (w/v) PEG 3350, 200 mmol/L ammonium citrate, pH 7.0) before frozen in liquid nitrogen.
Diffraction data were collected at the BL17U1 and BL19U1 beamlines of the Shanghai Synchrotron Radiation Facility, China (SSRF) and processed with autoPROC (Vonrhein et al., 2011). The apo structure was solved by molecular replacement using Phaser, with hTS (PDB: 1HVY) as the search model (McCoy et al., 2007). The complex structures were solved by molecular replacement using the refined apo structure as the search model. The geometry restraints of ligands were generated by the GRADE server (http://grade.globalphasing.org). Refinements of atomic coordinates, B factors and TLS parameters for each structure were done with autoBUSTER or Phenix. refine (Afonine et al., 2012; Smart et al., 2012). Alternately, models were manually optimized on Coot (Emsley et al., 2010). Data collection and refinement statistics are summarized in Supplementary Table S1. The coordinates and diffraction data of LvTS apo protein, LvTS-dUMP binary complex, LvTS-dUMP-raltitrexed ternary complex have been deposited in the PDB (www.rcsb.org) with accession numbers of 6K7Q, 6K7R and 6K7S, respectively.
The protein sequence alignments were performed with MEGA5.1 and ESPript 3.0 (Tamura et al., 2011; Robert and Gouet, 2014). Structural superimpositions were performed using Align (Satow et al., 1986). The Dali server was used to search for similar structures (Holm and Rosenström, 2010). Geometries were analyzed with MolProbity (Davis et al., 2007). All the structural images were generated using PyMOL (http://www.pymol.org).
3 RESULT AND DISCUSSION 3.1 Overall structures of apo LvTS, LvTS-dUMP and LvTS-dUMP-raltitrexedWe determined the crystal structure of apo LvTS at 2.27 Å resolution (Supplementary Table S1, Fig. 1a). The crystal belongs to space group P1, and one asymmetric unit contains 16 monomers. Two monomers are assembled as one dimer, in agreement with the result of size-exclusion chromatography (Supplementary Fig.S1b) and the previous reports (Stroud and Finer-Moore, 2003; Arvizu-Flores et al., 2009). Structures of all monomers are essentially identical, except for small differences in some flexible regions (Supplementary Fig.S2). Generally, loop23–27 (residues 23–27), segment88–102 (residues 88–102), and the C-terminal loop (residues 276–289) were not fully built due to undefined electron density, despite one exception that loop23–27 of monomer A is ordered. As expected, the overall structure of LvTS is similar to those of other known TSs. Each monomer consists of 7 α-helices and 8 β-strands, arranged in three layers: a bottom layer comprising a 6-stranded β-sheet, a interlayer formed by a long α-helix that is across the sheet and two shorter helices sitting aside the sheet, and a top layer containing four helices and two antiparallel β-strands. The 6-stranded β-sheets from two monomers stack against each other to form a dimer, and the interface contains hydrophobic and hydrophilic contacts between the sheets.
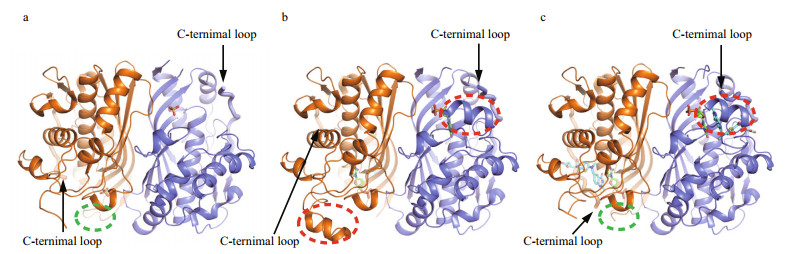
|
| Fig.1 Overall structures of LvTS a. apo form; b. LvTS-dUMP; c. LvTS-dUMP-raltitrexed. All the monomers A and B are colored as orange and slate, respectively. The ordered part of the C-terminal loop is bolded and indicated by arrow. The ordered loop23-27 and segment88-102 are indicated by green and red circles, respectively. |
The crystal structure of LvTS-dUMP was determined at 1.54 Å resolution (Supplementary Table S1, Fig. 1b). The crystal belongs to space group C 1 2 1, with one dimer per asymmetric unit. The overall structure is similar to the apo structure except for certain structural differences (Fig. 1a & b). Segment88–102 that is generally disordered in the apo structure is ordered in monomer B, containing a helical structural element. The same region of monomer A can also be modeled based on albeit weak electron density. The last eight residues of the C-terminal loop in monomer A are not involved in molecular interface and are disordered (Fig. 2a), while the corresponding residues in monomer B are largely ordered and partially cover the active site (Fig. 2b). As seen in the apo structure, loop23–27 is still disordered in both the monomers of LvTS-dUMP.
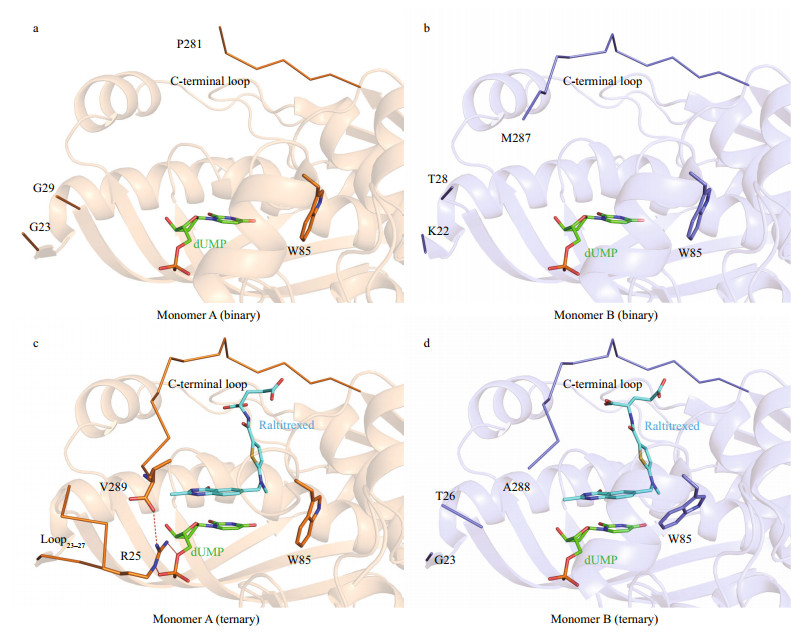
|
| Fig.2 Active-site region of LvTS a. monomer A of LvTS-dUMP; b. monomer B of LvTS-dUMP; c. monomer A of LvTS-dUMP-raltitrexed; d. monomer B of LvTS-dUMP-raltitrexed. The ligands and important residues are shown as sticks. The ordered part of the C-terminal loop and loop23-27 are shown as ribbon. Hydrogen bonds are shown as red dashed lines. |
The structure of LvTS-dUMP-raltitrexed was determined at 1.56 Å resolution (Supplementary Table S1, Fig. 1c). The crystal is isomorphous to that of LvTS-dUMP. Compared to LvTS-dUMP, LvTSdUMP-raltitrexed structure shows additional conformational changes, particularly for the C-terminal loop. In monomer A of LvTS-dUMPraltitrexed, the C-terminal loop is completely ordered to cap the active site; meanwhile, loop23–27 is ordered with a conformation different from that in monomer A of the apo form (Figs. 1a & c, 2c), and the guanidine group of Arg25 forms a salt bridge with the carboxyl group of Val289 (Fig. 2c). In monomer B of LvTSdUMP-raltitrexed, the last few residues of the C-terminal loop shift the position slightly to further cap the active site, while loop23–27 is still disordered (Fig. 2d). The C-terminal loops of the two monomers of LvTS-dUMP-raltitrexed can be well superposed (Supplementary Fig.S3), suggesting that the conformation observed here is not an artifact of the crystal packing. In addition, segment88–102 in monomer B is well ordered as seen in LvTS-dUMP, while the same region in monomer A can hardly be modeled due to very weak electron density. As the crystals of LvTS-dUMP and LvTS-dUMP-raltitrexed are isomorphous, the differences between the two structures largely reflect the effect of raltitrexed binding.
3.2 Interactions with ligands at the active siteBased on the high conservation of TSs, the active site of LvTS is supposed to be contributed by two monomers, comprising 15 residues (Arg25, Phe56, Gly59, Asn88, Tyr111, Cys171, His172, Arg191, Ser192, Leu197, Gly198, Phe201, Asn202, His232, Tyr234) from one monomer, and another two residues (Arg151 and Arg152) from the other monomer (Supplementary Fig.S4). Even in the absence of additional ligand, we were able to identify a sulfate ion bound to the active site of each monomer of apo LvTS (Fig. 3a). The sulfate ion is supposed to derive from the crystallization solution, and its binding has been also reported in previous TS structures (Schiffer et al., 1995; Deschamps et al., 2017). The sulfate ion makes several hydrogen bonds with the side chain atoms of Arg191 and Ser192 from one monomer, as well as the guanidine groups of Arg151 and Arg152 from the other.
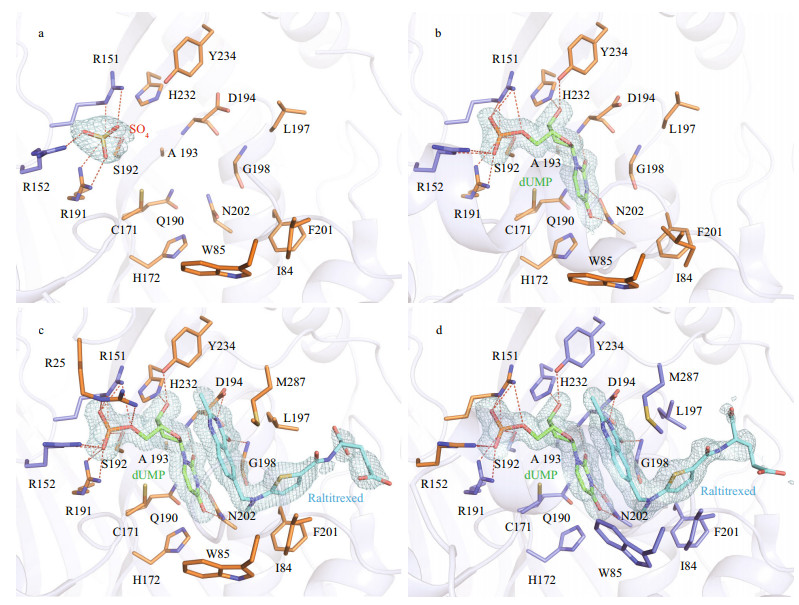
|
| Fig.3 Close-up of ligand-binding pocket a. apo form; b. LvTS-dUMP; c. monomer A of LvTS-dUMP-raltitrexed; d. monomer B of LvTS-dUMP-raltitrexed. Ion, ligands, and residues forming the ligand-binding pocket are shown as sticks. Hydrogen bonds are presented as red dashed lines. The weighted 2Fo-Fc electron density maps contoured at 1.0 σ level are shown as light blue mesh. |
In LvTS-dUMP (Fig. 3b), the uracil ring of dUMP is hydrogen bonded to the main chain amide of Asp194 and the side chain atoms of Asn202. The uracil ring also makes hydrophobic contacts with Cys171, His172, Gln190, and Ala193. C6 of dUMP is about 3.3 Å away from Sγ of the catalytic Cys171. The ribose ring of dUMP is hydrogen bonded to the side chain atoms of His232 and Tyr234. The phosphate moiety of dUMP is located at the position that corresponds to the sulfate ion, and hydrogen bonded to the same four residues that interact with the sulfate ion in the apo structure.
In LvTS-dUMP-raltitrexed (Fig. 3c & d), the interactions between dUMP and the protein retain almost identical to those in LvTS-dUMP, though the phosphate moiety of dUMP is additionally hydrogen bonded to the guanidine group of Arg25 in monomer A (Fig. 3c). The quinazoline ring of raltitrexed is stacked against the uridine ring of dUMP, and further stabilized by the hydrogen bonds with the carboxyl group of Asp194 and the main chain NH group of Gly198. Moreover, the quinazoline makes a hydrophobic interaction with Trp85 in monomer B (Fig. 3d), but not in monomer A. The thiophene ring of raltitrexed is surrounded by four hydrophobic residues Ile84, Leu197, Phe201, and Met287. The glutamate tail of raltitrexed only showed weak electron density, without direct interaction with surrounding residues. We modeled two conformations of this moiety in the two monomers to better interpret the electron density (Fig. 3c & d).
3.3 Comparison with other known TSsThe structural similarity searching server Dali (Holm and Rosenström, 2010) identified hTS and mTS as the top 2 hits based on 3D similarity, both with the root-mean-square deviation for the aligned Cα of 0.5–0.6 Å (Supplementary Fig.S5). LvTS shares 75.96% and 75.69% sequence identities with hTS and mTS, respectively. Accordingly, we mainly compared LvTS-dUMP-raltitrexed complex with those of hTS and mTS to show species-specific differences, focusing on the ligand-enzyme interactions.
All the known TS-dUMP-raltitrexed complexes primarily adopt either the open or the closed conformation. In the open conformation, the C-terminal loop is disordered and fails to cap the active site; C6 of dUMP is 2.7–3.5 Å away from Sγ of the catalytic cysteine (Carreras and Santi, 1995; Dowierciał et al., 2014). In the closed conformation, the C-terminal loop is ordered to cap the active site cavity, accompanied by inward movement of many other secondary structural elements; typically, C6 of dUMP and Sγ of the catalytic cysteine lie in a short distance of 2.4–2.5 Å (in dimer AB of mTS, PDB: 4EB4) or even form a covalent bond (in hTS, PDB: 1HVY) (Phan et al., 2001; Dowierciał et al., 2014). Notably, one monomer of hTS-dUMP-raltitrexed (PDB: 5X5Q) exhibits an atypical closed conformation when the complex crystals were prepared using the soaking method: raltitrexed binding could induce ordering of the C-terminal loop, but the covalent bond is not formed between C6 of dUMP and Sγ of the catalytic cysteine with the distance between the two atoms similar to that in the apo and the binary structures (Chen et al., 2017). The covalent-bond formation might be hindered due to the short soaking time or the crystal lattice restraints.
LvTS-dUMP-raltitrexed adopts a closed conformation, but it shows structural features obviously different from hTS (PDB: 1HVY) and mTS (dimer AB, PDB: 4EB4) in the closed conformation. The C-terminal loop of LvTS shows a position shift away from the substrate-binding pocket, compared to those of hTS and mTS (Fig. 4a). The sequences of the C-terminal loops of LvTS, hTS and mTS share a high similarity, but Lys282 of LvTS substitutes for the threonine in the corresponding positions of hTS and mTS (Fig. 4a). Substitution into a residue with a long side chain may interfere with interactions with raltitrexed, which may result in the position shift of the C-terminal loop in LvTS-dUMP-raltitrexed. Interestingly, the distances between C6 of dUMP and Sγ of the catalytic Cys171 in LvTS-dUMP-raltitrexed are 3.2 Å in monomer A and 3.0 Å in monomer B (Fig. 4b). The distances are obviously longer than those in the known TS structures with closed conformations, but are similar to those in the TS structures with open conformations. In monomer B of LvTS-dUMP-raltitrexed, raltitrexed sits in a similar position to that in hTS and mTS; Trp85 tilts the indole ring to make hydrophobic contact with raltitrexed, resembling the corresponding residues in hTS (Trp109) and mTS (Trp103) (Fig. 4a). In monomer A, raltitrexed showed a position deviation from that in hTS and mTS, and the orientation of the indole ring of Trp85 is similar to that in the apo and binary structures that presents the open conformation (Figs. 2 & 4a). Besides, loop23–27 becomes ordered only in monomer A of LvTS-dUMP-raltitrexed, where Arg25 makes contact with dUMP (Fig. 2c). This is different from that in hTS and mTS, where the corresponding loops are ordered in both monomers to interact with dUMP. Taken together, these findings indicate that LvTSdUMP-raltitrexed adopts a loosely closed conformation, with structural features intermediate between the closed and the open conformations reported previously. These species-specific variations at the active-site regions are valuable in designing competitive inhibitors.
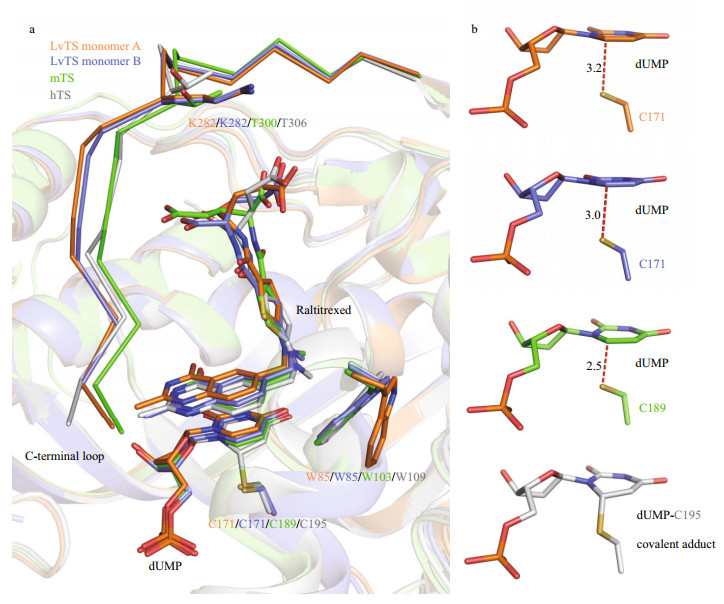
|
| Fig.4 Structural comparison of TS-dUMP-raltitrexed complexes a. superimposition of active-site regions of LvTS monomer A and B, mTS (dimer AB, PDB: 4EB4), and hTS (PDB: 1HVY). The ligands and residues are shown as sticks. The C-terminal loops are shown as ribbon; b. interactions between C6 of dUMP and Sγ of the catalytic cysteine in LvTS monomer A and B, mTS (dimer AB, PDB: 4EB4), and hTS (PDB: 1HVY). The distances (Å) between the two atoms in LvTS and mTS are indicated. |
In addition, TS contains an allosteric regulation site that could be used for designing allosteric inhibitors. This site is located near the dimer interface and close to the active-site loop (Cardinale et al., 2011). Previous studies showed that the active-site loop (residues 181–196) of hTS can adopt two conformations, the inactive and the active. The inactive conformation can be stabilized by Arg163 of hTS (Gibson et al., 2008). However, other TSs, including LvTS, lack such an arginine residue, and their active-site loops only exhibit the active conformation. Nevertheless, a peptide ligand can also stabilize the inactive conformation of the active-site loop when bound to the allosteric site of hTS (PDB: 3N5E) (Supplementary Fig.S4) (Cardinale et al., 2011). This raises a possibility to design allosteric inhibitors for other TSs that lack hTS-Arg163 counterpart. The residues that interact with the allosteric peptide in hTS are largely identical to those in LvTS, except that Arg163 of hTS is replaced by Gln139 in LvTS (Supplementary Fig.S4), implying a potential variation in ligand-binding property. As these residues of the allosteric site are not essential for catalysis, we proposed that this region may exhibit more species-specific variations among TSs of shrimp and its pathogens, which would make allostericinhibitor optimization easier to gain a high species selectivity.
4 CONCLUSIONHerein, we presented the shrimp TS crystal structures in the apo and ligand-bound states. This is the first structural report on crustacean TS. From the accurate structural information, we were able to observe the species-specific differences between LvTS and other close homologs. In the future, with solution of shrimp-pathogen TS structures, we expect to identify species-specific differences between LvTS and pathogen TSs to design pathogen-specific inhibitors.
5 DATA AVAILABILITY STATEMENTThe data that support the findings of this study are available on request from the corresponding author.
6 ACKNOWLEDGMENTWe thank the staffs from the BL17U1 and BL19U1 beamline stations at SSRF for assistance during data collection.
Electronic supplementary materialSupplementary material (Supplementary Table S1 and Figs.S1–S5) is available in the online version of this article at https://doi.org/10.1007/s00343-019-9184-8.
Afonine P V, Grosse-Kunstleve R W, Echols N, Headd J J, Moriarty N W, Mustyakimov M, Terwilliger T C, Urzhumtsev A, Zwart P H, Adams P D. 2012. Towards automated crystallographic structure refinement with Phenix.refine. Acta Crystallographica Section D:Structural Biology, 68(4): 352-367.
DOI:10.1107/S0907444912001308 |
Arvizu-Flores A A, Aispuro-Hernandez E, Garcia-Orozco K D, Varela-Romero A, Valenzuela-Soto E, VelazquezContreras E F, Rojo-Domínguez A, Yepiz-Plascencia G, Maley F, Sotelo-Mundo R R. 2009. Functional identity of the active sites of crustacean and viral thymidylate synthases. Comparative Biochemistry and Physiology Part C:Toxicology & Pharmacology, 150(3): 406-413.
DOI:10.1016/j.cbpc.2009.06.008 |
Aslanidis C, de Jong P J. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Research, 18(20): 6 069-6 074.
DOI:10.1093/nar/18.20.6069 |
Cardinale D, Guaitoli G, Tondi D, Luciani R, Henrich S, SaloAhen O M H, Ferrari S, Marverti G, Guerrieri D, Ligabue A, Frassineti C, Pozzi C, Mangani S, Fessas D, Guerrini R, Ponterini G, Wade R C, Costi M P. 2011. Proteinprotein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase. Proceedings of the National Academy of Sciences of the United States of America, 108(34): E542-E549.
DOI:10.1073/pnas.1104829108 |
Carreras C W, Santi D V. 1995. The catalytic mechanism and structure of thymidylate synthase. Annual Review of Biochemistry, 64: 721-762.
DOI:10.1146/annurev.bi.64.070195.003445 |
Chen D, Jansson A, Sim D, Larsson A, Nordlund P. 2017. Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states. The Journal of Biological Chemistry, 292(32): 13 449-13 458.
DOI:10.1074/jbc.M117.787267 |
Choi Y M, Yeo H K, Park Y W, Lee J Y. 2016. Structural analysis of thymidylate synthase from Kaposi's sarcomaassociated herpesvirus with the anticancer drug raltitrexed. PLoS One, 11(12): e0168019.
DOI:10.1371/journal.pone.0168019 |
Davis I W, Leaver-Fay A, Chen V B, Block J N, Kapral G J, Wang X Y, Murray L W, Arendall III W B, Snoeyink J, Richardson J S, Richardson D C. 2007. MolProbity:allatom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Research, 35(S2): W375-W383.
DOI:10.1093/nar/gkm216 |
de Clercq E, Balzarini J, Descamps J, Bigge C F, Chang C T C, Kalaritis P, Mertes M P. 1981. Antiviral, antitumor, and thymidylate synthetase inhibition studies of 5-substituted styryl derivatives of 2'-deoxyuridine and their 5'-phosphates. Biochemical Pharmacology, 30(5): 495-502.
DOI:10.1016/0006-2952(81)90635-3 |
Deschamps P, Réty S, Bareille J, Leulliot N. 2017. Crystal structure of the active form of native human thymidylate synthase in the absence of bound substrates. Acta Crystallographica Section F:Structural Biology Communications, 73(6): 336-341.
DOI:10.1107/S2053230X17007233 |
Dowierciał A, Wilk P, Rypniewski W, Rode W, Jarmuła A. 2014. Crystal structure of mouse thymidylate synthase in tertiary complex with dUMP and raltitrexed reveals N-terminus architecture and two different active site conformations. Biomed Research International, 2014: 945803.
DOI:10.1155/2014/945803 |
Emsley P, Lohkamp B, Scott W G, Cowtan K. 2010. Features and development of Coot. Acta Crystallographica Section D:Structural Biology, 66(4): 486-501.
DOI:10.1107/S0907444910007493 |
Flegel T W. 2012. Historic emergence, impact and current status of shrimp pathogens in Asia. Journal of Invertebrate Pathology, 110(2): 166-173.
DOI:10.1016/j.jip.2012.03.004 |
Food and Agriculture Organization of the United Nations Globefish. 2018. Market Reports, http://www.fao.org/inaction/globefish.
|
Gibson L M, Lovelace L L, Lebioda L. 2008. The R163K mutant of human thymidylate synthase is stabilized in an active conformation:structural asymmetry and reactivity of cysteine 195. Biochemistry, 47(16): 4 636-4 643.
DOI:10.1021/bi7019386 |
Holm L, Rosenström P. 2010. Dali server:conservation mapping in 3D. Nucleic Acids Research, 38(S2): W545-W549.
DOI:10.1093/nar/gkq366 |
Jackman A L. 1999. Antifolate Drugs in Cancer Therapy. Humana Press, Totowa NJ. 456p, https://doi.org/10.1007/978-1-59259-725-3.
|
Lightner D V, Redman R M, Pantoja C R, Tang K F J, Noble B L, Schofield P, Mohney L L, Nunan L M, Navarro S A. 2012. Historic emergence, impact and current status of shrimp pathogens in the Americas. Journal of Invertebrate Pathology, 110(2): 174-183.
DOI:10.1016/j.jip.2012.03.006 |
McCoy A J, Grosse-Kunstleve R W, Adams P D, Winn M D, Storoni L C, Read R J. 2007. Phaser crystallographic software. Journal of Applied Crystallography, 40(4): 658-674.
DOI:10.1107/S0021889807021206 |
Perry K M, Fauman E B, Finer-Moore J S, Montfort W R, Maley G F, Maley F, Stroud R M. 1990. Plastic adaptation toward mutations in proteins:structural comparison of thymidylate synthases. Proteins, 8(4): 315-333.
DOI:10.1002/prot.340080406 |
Phan J, Koli S, Minor W, Dunlap R B, Berger S H, Lebioda L. 2001. Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug. Biochemistry, 40(7): 1 897-1 902.
DOI:10.1021/bi002413i |
Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research, 42(W1): W320-W324.
DOI:10.1093/nar/gku316 |
Rustum Y M, Harstrick A, Cao S, Vanhoefer U, Yin M B, Wilke H, Seeber S. 1997. Thymidylate synthase inhibitors in cancer therapy:direct and indirect inhibitors. Journal of Clinical Oncology, 15(1): 389-400.
DOI:10.1200/JCO.1997.15.1.389 |
Satow Y, Cohen G H, Padlan E A, Davies D R. 1986. Phosphocholine binding immunoglobulin Fab McPC603:an X-ray diffraction study at 2. 7 Å. Journal of Molecular Biology, 190(4): 593-604.
DOI:10.1016/0022-2836(86)90245-7 |
Schiffer C A, Clifton I J, Davisson V J, Santi D V, Stroud R M. 1995. Crystal structure of human thymidylate synthase:a structural mechanism for guiding substrates into the active site. Biochemistry, 34(50): 16 279-16 287.
DOI:10.1021/bi00050a007 |
Smart O S, Womack T O, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. 2012. Exploiting structure similarity in refinement:automated NCS and target-structure restraints in BUSTER. Acta Crystallographica Section D:Structural Biology, 68(4): 368-380.
DOI:10.1107/S0907444911056058 |
Stout T J, Tondi D, Rinaldi M, Barlocco D, Pecorari P, Santi D V, Kuntz I D, Stroud R M, Shoichet B K, Costi M P. 1999. Structure-based design of inhibitors specific for bacterial thymidylate synthase. Biochemistry, 38(5): 1 607-1 617.
DOI:10.1021/bi9815896 |
Stroud R M, Finer-Moore J S. 2003. Conformational dynamics along an enzymatic reaction pathway:thymidylate synthase, "the movie". Biochemistry, 42(2): 239-247.
DOI:10.1021/bi020598i |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5:molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10): 2 731-2 739.
DOI:10.1093/molbev/msr121 |
Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. 2011. Data processing and analysis with the autoPROC toolbox. Acta Crystallographica Section D:Structural Biology, 67(4): 293-302.
DOI:10.1107/S0907444911007773 |
Zaware N, Sharma H, Yang J, Devambatla R K V, Queener S F, Anderson K S, Gangjee A. 2013. Discovery of potent and selective inhibitors of Toxoplasma gondii thymidylate synthase for opportunistic infections. ACS Medicinal Chemistry Letters, 4(12): 1 148-1 151.
DOI:10.1021/ml400208v |
 2020, Vol. 38
2020, Vol. 38


