Shanghai University
Article Information
- WANG Qian (王倩), TAN Xungang (谭训刚), DU Shaojun (杜少军), SUN Wei (孙威) , YOU Feng (尤锋), ZHANG Peijun (张培军)
- Characterization, tissue distribution, and expression of neuropeptide Y in olive flounder Paralichthys olivaceus
- Chinese Journal of Oceanology and Limnology, 2015, 33(3): 553-558
- http://dx.doi.org/10.1007/s00343-015-4090-1
Article History
- Received Apr. 4, 2014; ;
- accepted in principle May 14, 2014
- accepted for publication Nov. 4, 2014
2 University of Chinese Academy of Sciences, Beijing 100049, China;
3 Department of Biochemistry and Molecular Biology, School of Medicine, University of Maryland 701 E. Pratt St, Baltimore MD, 21202, USA
Neuropeptide Y(NPY)was originally recognized in porcine brain(Tatemoto, 1982), and belongs to the neuropeptide Y family that includes tetrapod pancreatic polypeptide(PP), pancreatic polypeptide Y(PY), and peptide YY(PYY)(Cerda-Reverter et al., 2000). The 36-amino acid peptide NPY is produced from a 96 aa pre-pro-peptide that contains a 28 aa N-terminal signal peptide and a 32 aa C-terminal extension. NPY is highly conserved in vertebrates(Kurokawa and Suzuki, 2002).
NPY mainly expresses in the brain. Its transcripts can also be expressed in peripheral tissues such as the cardiovascular system, adrenal gland, and gastrointestinal tract(Dumont et al., 1992). As a signaling molecule, it plays key roles in regulating a variety of physiological processes including food intake, circadian rhythms, neuroendocrine functions, and glucose homeostasis(Larhammar, 1996; Zhang, 2012; Tang, 2013). It has been reported that NPY in fish stimulates the secretion of gonadotropin-II(GTHII), growth hormone(GH), and luteinizing hormone(LH)(Peng et al., 1993; Cerda-Reverter et al., 1999). Intracerebro-ventricular(ICV)injection of NPY stimulates food intake in goldfish, rainbow trout, and tilapia(Lopez-Patino et al., 1999; Aldegunde and Mancebo, 2006; Kehoe and Volkoff, 2007).Additionally, intraperitoneal injection of NPY dose-dependently stimulates tilapia growth(Carpio et al., 2006). Moreover, dietary restriction and food deprivation significantly increase brain NPY gene expression in both goldfish(Narnaware et al., 2000) and zebrafish(Yokobori et al., 2012). NPY is thus an important factor that regulates feeding behavior and may be a key orexigenic neuropeptide.
The olive flounder(Paralichthys olivaceus)is an economically important marine fish species and is widely cultured in northern China. Growth rate and weight gain are important factors for flounder aquaculture. NPY is a potentially useful regulator of fish feeding and growth in this sector. In the present study, we characterized the tissue-specific NPY expression and examined changes in its expression around feeding time and during food deprivation. Our results demonstrate that NPY mRNA levels increase prior to food intake and subsequently decrease in the entire olive fl ounder brain. Moreover, food deprivation for 48 h reduced NPY expression levels. Therefore, NPY expression is associated with food intake and may have a potential application in olive flounder aquaculture. 2 MATERIAL AND METHOD2.1 Animal
Olive flounder(11.67±1.14 cm, 12.06±2.84 g)were originally obtained from Nanshan market(Qingdao, China) and cultured in a 500-L tank under laboratory conditions. The fish were maintained at 15.0±1.0°C and fed with commercial fish food(Shengsuo, Sh and ong, China)containing 47% crude protein and 8% crude lipid. The fish were fed at a fixed time of 15:00 PM once a day for 20 days prior to the experiments. For sampling, the fish were anesthetized with 100 mg/L MS222(tricaine methanesulphonate) and euthanized by spinal section. Three fish were used to analyze NPY tissue distribution. The whole brain, testes, ovary, heart, liver, intestine, stomach, head kidney, muscle, kidney, spleen, gill, and retina were sampled(n =3). To investigate NPY expression during feeding time and food deprivation, the flounder were divided into two tanks a day before the experiment. On the experiment day, fish in one tank were fed regularly and sampled around feeding time; fish in the other tank were not fed at the scheduled time(0 h) and fasted for 48 h. The entire brain was dissected. Samples were frozen immediately in liquid nitrogen and stored at -80°C. 2.2 Isolation of total RNA and reverse transcriptionPCR
Total RNA was extracted with Trizol reagent(Invitrogen, USA)following the manufacturer's instructions. The RNA concentration was determined by measuring the absorbance at 260 nm with a NanoDrop-1000 spectrophotometer(Thermo Fisher Scientific, USA). After digestion with DNase I(TaKaRa, Japan), cDNA was synthesized with M-MLV Reverse Transcriptase(Promega, USA)using 2 μg RNA as a template.
The NPY RT-PCR( and qRT-PCR)primers were designed according to GenBank sequence No. AB055211.1. The internal control PCR reactions were performed using the housekeeping gene β-actin and the β-actin-F/R primers were designed according to GenBank sequence No. HQ386788.1(Table 1). PCR amplifications were set up in a 25-μL volume containing 1 μL cDNA template, 0.4 μmol/L each forward and reverse primer, and 12.5 μL 2× Es Taq MasterMix(CoWin Biotech, Beijing, China). The PCR parameters were as follows: 5 min initial denaturation at 94°C, then 35 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 1 min, and finally 10 min at 72°C.
The NPY mRNA levels during feeding time and food deprivation were detected by qRT-PCR. The primers for qRT-PCR were designed by TaKaRa(Japan)(NPY-qF and NPY-qR, Table 1). The internal control PCR reactions were performed using β-actin(β-actin-qF and β-actin-qR, Table 1). Initial validation experiments were conducted to ensure the equivalent PCR efficiencies of both primers. The qRT-PCR was carried out on a TaKaRa Thermal Cycler Dice™ Real Time System TP800 machine using SYBR® Premix Ex Taq™ II(Tli RNaseH Plus)(TaKaRa, Japan). The qRT-PCR conditions were as follows: 30 s at 95°C, then 40 cycles of 5 s at 95°C, 30 s at 55°C and 20 s at 72°C. A melting curve analysis was also added at the end of each qRT-PCR reaction to ensure the specificity of the amplification. Each PCR run was conducted in triplicate. The relative gene expression was analyzed by the 2-∆∆Ctmethod(Livak and Schmittgen, 2001). 2.4 Phylogenetic analysis of flounder NPY
NPY coding sequences of different species were aligned in ClustalX Multiple alignment software 1.83 and the phylogenetic tree was constructed in MEGA5 by the maximum likelihood method(Jones et al., 1992; Tamura et al., 2011). Xenopus laevis was used as an outgroup. The bootstrap method with 1 000 replications was applied to test the phylogeny. 2.5 NPY gene expression at different feeding times
Flounder were fed daily at the fixed time(15:00 PM). On the experimental day, they were r and omly sampled at a set time: 3 h prior to feeding(-3 h group), 1 h prior to feeding(-1 h group), at feeding time(0 h group), 1 h after feeding(1 h group), and 3 h after feeding(3 h group). Fish in the 0 h group were allowed to feed for 5 min at the scheduled feeding time and subsequently sampled. Each group contained three fish. All samples were euthanized and the brains were isolated. 2.6 Fasting treatments
To evaluate the effect of fasting, feeding was stopped at the scheduled time and the fish were deprived of food for the next 48 h. Fish were sampled at 24 h(24 h fasted group) and 48 h(48 h fasted group). Each group contained three fish. The 0 h group treated above was used as a control in this fasting experiment. All samples were euthanized and the brains were dissected. 2.7 Data analysis
Relative NPY mRNA levels are presented as mean±SEM. Differences among groups were analyzed by one-way ANOVA followed by Tukey’s tests, using GraphPad Prism Version 5.01 software(San Diego, CA, USA). Statistically significant differences were defined as P<0.05. 3 RESULT3.1 Isolation of olive flounder NPY cDNA and phylogenetic analysis
A 369-bp fragment for olive flounder NPY was cloned according to the GenBank sequence AB055211.1. As described in a previous study, the fragment contained a 300-bp open reading frame(ORF), which was composed of a signal peptide of 28 amino acids, a mature 36 amino acid peptide(containing the Gly-Lys-Arg processing site), and a 32 amino acid carboxy-terminal extension(Kurokawa and Suzuki, 2002). In the phylogenetic tree(Fig. 1)NPY in olive flounder and other bony fish, e.g., P. americanus, P. orbignyanus, E. coioides, O. niloticus, and O. latipes, clustered together, while that in D. rerio, C. auratus, and C. gibelioformed another cluster. L. ocellata, X. laevis, G. gallus, and mammalian NPY formed separate clusters.
 |
|
Fig. 1 Phylogenetic tree of NPY from Paralichthys olivaceus(AB055211.1), Pseudopleuronectes americanus(EU684053.1), Paralichthys orbignyanus(FJ705358.1), Oreochromis niloticus(XM_003448854.2), Epinephelus coioides(AY626561.1), Oryzias latipes(EU047761.1), Siniperca chuatsi(EF554594.1), Dicentrarchus labrax(AJ005378.2), Leucopsarionpetersii(AB733443.1), Gadus morhua(DQ256082.1), Oncorhynchus mykiss(NM_001124266.1), Salmo salar(AB455539.1), Danio rerio(NM_131074.2), Carassius auratus(M87297.1), Carassius gibelio(JN885181.1), Leucorajaocellata(EU684052.1), Xenopus laevis(L07413.1), Gallus gallus(NM_205473.1), Homo sapiens(NM_000905.3), and Rattus norvegicus(NM_012614.2)were used as an outgroup The tree was built using the maximum likelihood method, bootstrapped with 1 000 replicates. |
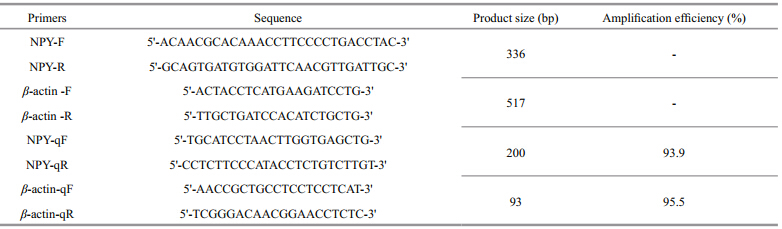
To investigate NPY expression in various olive flounder tissues, RT-PCR was used to analyze NPY mRNA levels in 13 tissues(Fig. 2a). Strong expression was observed in the brain, and moderate amplification signals were found in the retina and gill. Low levels of expression were detected in the testes, ovary, heart, head kidney, kidney, and spleen, while no NPY expression was detected in the liver, intestine, stomach, or muscle. The levels of β-actin transcripts were similar among the 13 different tissues(Fig. 2b).
 |
|
Fig. 2 NPY expression in different olive flounder tissues a. PCR products from NPY transcripts; b. PCR products of β-actin RNA transcripts. Ladder: 100 bp marker; T: testis; O: ovary; H: heart; B: brain; L: liver; I: intestine; St: stomach; Hk: head kidney; M: muscle; K: kidney; Sp: spleen; G: gill; R: retina; NC: negative control. |
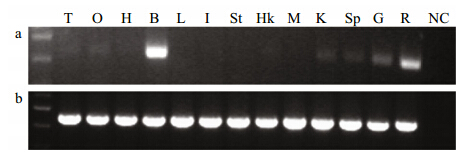
To evaluate the effect of feeding schedule on NPY expression, we analyzed NPY gene expression in the fish brain around the fixed feeding time. A trend of increasing before and decreasing after feeding time(15:00 PM)was observed. Compared with the -1 h group before feeding, NPY expression in the 3 h group significantly decreased after feeding(P<0.05)(Fig. 3).
 |
|
Fig. 3 NPY gene expression around feeding time in olive flounder Data are presented as means±SEM(n =3). Different letters above the bars represent statistically significant differences(P<0.05). |
To determine whether food deprivation had any effect on NPY expression, we compared the levels of NPY mRNA expression in the fish brains between the fed and unfed groups. Compared with the fish fed at the scheduled feeding time(0 h), NPY expression decreased significantly(P<0.05)in both the 24 h and 48 h fasted groups. In the 24 h fasted group, NPY mRNA levels decreased by 81.7%, while in the 48 h fasted group they decreased approximately by 91.7%(Fig. 4).
 |
|
Fig. 4 Effects of food deprivation on NPY gene expression Data are presented as means±SEM(n =3). Different letters above the bars represent statistically significant differences(P<0.05). |
To investigate the role of NPY during food intake in olive flounder, the NPY cDNA was cloned according to GenBank sequence AB055211.1. Its distribution pattern in different tissues and the effect of feeding and fasting on expression were analyzed.
The NPY contained a 300-bp ORF and the deduced amino acid sequence was conserved with many other fish and mammals. The phylogenetic analysis demonstrated that olive flounder NPY exhibited high identity with that of bony fish. The mRNA distribution was analyzed by RT-PCR in various tissues. The results revealed that NPY was highly expressed in the brain and retina, but rarely in the digestive organs. These findings were consistent with previous studies on Japanese flounder(Kurokawa and Suzuki, 2002) and Atlantic salmon(Murashita et al., 2009). The conserved coding sequence and tissue distribution suggested similar physiological roles of NPY in flounder and other organisms.
In our present study, NPY expression levels in the brain exhibited an increase prior to and a decrease after food intake. NPY expression decreased significantly in the 3 h group after feeding compared with that in the -1 h group before feeding(P<0.05). This result is in agreement with a previous study on goldfish(Narnaware et al., 2000). Additionally, increased NPY levels have also been reported in Atlantic cod forebrains during meal times with a decline 2 h after that(Kehoe and Volkoff, 2007). We presume that the pre-pr and ial increases in NPY mRNA expression before the scheduled feeding time indicate the sensation of hunger or the anticipation of food in flounder. Thus, the gradual decrease in NPY mRNA levels may be explained by the acquisition of food. Taken together, our results support the hypothesis that NPY stimulates feeding as a “hunger signal” and may be involved in regulating food intake in olive flounder.
In the fasting experiment, fish in the 24 h and 48 h fasted groups showed an ~81.7% and 91.7% decrease in NPY mRNA levels, respectively, compared with those in the fed group(0 h). However, NPY expression exhibited different changes in other fish species. In goldfish, Brazilian flounder, and winter skate, 72 h food deprivation led to a significant and time-related increase in NPY expression levels(Narnaware et al., 2000; MacDonald and Volkoff, 2009; Campos et al., 2010). Food deprivation had no significant effect on NPY expression in either zebrafish(Drew et al., 2008)or Atlantic salmon(Murashita et al., 2009). In other reports, NPY expression decreased in the winter skate hypothalamus after 2 weeks of fasting(MacDonald and Volkoff, 2009) and in the forebrain of Atlantic cod fasted for 7 days(Kehoe and Volkoff, 2007). Therefore, NPY expression in olive flounder observed in this study may be for the following reasons:(1)different species may display different responses. Olive flounder are naturally docile, have low locomotion activity, and are not easily agitated. These characteristics may lead to a relatively slow rate of energy consumption;(2)NPY has different expression patterns in different regions of the brain. In this study, we used the entire brain to ascertain the relationship between NPY and food intake. This may be one explanation for the differences between our report and others;(3)olive flounder is a cold-temperate species with a relatively low metabolism under low temperatures. They may have a higher tolerance for periods of starvation than other species. Therefore, the decrease in NPY expression levels in our food deprivation experiment may be a hunger stress response in olive flounder. 5 CONCLUSION
In summary, NPY distribution in olive flounder tissues was analyzed and the relationship between NPY expression levels and food intake was investigated. NPY expression increased shortly before feeding time and decreased after fasting for 2 days. These results indicate the function of NPY in stimulating food intake and its potential importance in olive flounder aquaculture.
| Aldegunde M, Mancebo M. 2006. Effects of neuropeptide Y on food intake and brain biogenic amines in the rainbow trout (Oncorhynchus mykiss). Peptides, 27 (4): 719-727. |
| Campos V F, Collares T, Deschamps J C, Seixas F K,Dellagostin O A, Lanes C F C, Sandrini J, Marins L F,Okamoto M, Sampaio L A, Robaldo R B. 2010.Identification, tissue distribution and evaluation of brain neuropeptide Y gene expression in the Brazilian flounder Paralichthys orbignyanus. J. Biosci., 35 (3): 405-413. |
| Carpio Y, Acosta J, Morales A, Herrera F, Gonzalez L J,Estrada M P. 2006. Cloning, expression and growth promoting action of Red tilapia (Oreochromis sp.) neuropeptide Y. Peptides, 27 (4): 710-718. |
| Cerda-Reverter J M, Martinez-Rodriguez G, Zanuy S, Carrillo M, Larhammar D. 2000. Molecular evolution of the neuropeptide Y (NPY) family of peptides: cloning of three NPY-related peptides from the sea bass (Dicentrarchus labrax). Regul. Pept., 95 (1-3): 25-34. |
| Cerda-Reverter J M, Sorbera L A, Carrillo M, Zanuy S. 1999.Energetic dependence of NPY-induced LH secretion in a teleost fish (Dicentrarchus labrax). Am. J. Physiol. Regul.Integr. Comp. Physiol., 277 (6): R1627-R1634. |
| Drew R E, Rodnick K J, Settles M, Wacyk J, Churchill E,Powell M S, Hardy R W, Murdoch G K, Hill R A, Robison B D. 2008. Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio). Physiol.Genomics, 35 (3): 283-295. |
| Dumont Y, Martel J C, Fournier A, Stpierre S, Quirion R. 1992.Neuropeptide-Y and neuropeptide-Y receptor subtypes in brain and peripheral-tissues. Prog. Neurobiol., 38 (2): 125-167. |
| Jones D T, Taylor W R, Thornton J M. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci., 8 : 275-282. |
| Kehoe A S, Volkoff H. 2007. Cloning and characterization of neuropeptide Y (NPY) and cocaine and amphetamine regulated transcript (CART) in Atlantic cod (Gadus morhua). Comp. Biochem. Physiol. A-Mol. Integr.Physiol., 146 (3): 451-461. |
| Kiris G A, Kumlu M, Dikel S. 2007. Stimulatory effects of neuropeptide Y on food intake and growth of Oreochromis niloticus. Aquaculture, 264 (1-4): 383-389. |
| Kurokawa T, Suzuki T. 2002. Development of neuropeptide Y-related peptides in the digestive organs during the larval stage of Japanese flounder, Paralichthys olivaceus. Gen.Comp. Endocrinol., 126 (1): 30-38. |
| Larhammar D. 1996. Evolution of neuropeptide Y, peptide YY and pancreatic polypeptide. Regul. Pept., 62 (1): 1-11. |
| Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Method s, 25 (4): 402-408. |
| Lopez-Patino M A, Guijarro A I, Isorna E, Delgado M J,Alonso-Bedate M, de Pedro N. 1999. Neuropeptide Y has a stimulatory action on feeding behavior in goldfish (Carassius auratus). Eur. J. Pharmacol., 377 (2-3): 147-153. |
| MacDonald E, Volkoff H. 2009. Neuropeptide Y (NPY), cocaine-and amphetamine-regulated transcript (CART) and cholecystokinin (CCK) in winter skate (Raja ocellata): cDNA cloning, tissue distribution and mRNA expression responses to fasting. Gen. Comp. Endocrinol., 161 (2): 252-261. |
| Murashita K, Kurokawa T, Ebbesson L O E, Stefansson S O,Ronnestad I. 2009. Characterization, tissue distribution, and regulation of agouti-related protein (AgRP), cocaineand amphetamine-regulated transcript (CART) and neuropeptide Y (NPY) in Atlantic salmon (Salmo salar).Gen. Comp. Endocrinol., 162 (2): 160-171. |
| Narnaware Y K, Peyon P P, Lin X W, Peter R E. 2000.Regulation of food intake by neuropeptide Y in goldfish.Am. J. Physiol.-Regul. Integr. Comp. Physiol., 279 (3):R1025-R1034. |
| Peng C, Chang J P, Yu K L, Wong A O L, Vangoor F, Peter R E, Rivier J E. 1993. Neuropeptide-Y stimulates growthhormone and gonadotropin-ii secretion in the goldfish pituitary: involvement of both presynaptic and pituitary cell actions. Endocrinology, 132 (4): 1 820-1 829. |
| Silverstein J T, Plisetskaya E M. 2000. The effects of NPY and insulin on food intake regulation in fish. Am. Zool., 40 (2): 296-308. |
| Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol.Evol., 28 (10): 2 731-2 739. |
| Tang Z G, Sun C Y, Yan A F, Wu S G, Qin C B, Zhang Y H, Li W S. 2013. Genes involved in fatty acid metabolism: molecular characterization and hypothalamic mRNA response to energy status and neuropeptide Y treatment in the orange-spotted grouper Epinephelus coioides. Mol Cell Endocrinol., 376 (1-2): 114-124. |
| Tatemoto K. 1982. Neuropeptide-Y: complete amino-acidsequence of the brain peptide. Proc. Natl. Acad. Sci. USA, 79 (18): 5 485-5 489. |
| Yokobori E, Azuma M, Nishiguchi R, Kang K S, Kamijo M,Uchiyama M, Matsuda K. 2012. Neuropeptide Y stimulates food intake in the zebrafish, Danio rerio. J.Neuroendocrinol., 24 (5): 766-773. |
| Zhang L, Nguyen A D, Lee I-C J, Yulyaningsih E, Riepler S J,Stehrer B, Enriquez R F, Lin S, Shi Y-C, Baldock P A,Sainsbury A, Herzog H. 2012. NPY modulates PYY function in the regulation of energy balance and glucose homeostasis. Diabetes Obes Metab., 14 (8): 727-736. |
 2015, Vol. 33
2015, Vol. 33