Shanghai University
Article Information
- FANG Yalu (方亚璐), KAI Yoshiaki, YANAGIMOTO Takashi, SONG Na (宋娜), GAO Tianxiang (高天翔)
- A new record of Sebastes koreanus from China based on morphological characters and DNA barcoding
- Chinese Journal of Oceanology and Limnology, 2015, 33(3): 590-596
- http://dx.doi.org/10.1007/s00343-015-4166-y
Article History
- Received Jun. 6, 2014
- accepted in principle Aug. 28, 2014;
- accepted for publication Oct. 23, 2014
2 Maizuru Fisheries Research Station, Field Science Education and Research Centre, Kyoto University, Kyoto 625-0086, Japan;
3 National Research Institute of Fisheries Science,;
2-12-4, Yokohama, 236-8648, Japan;
4 Fishery College, Zhejiang Ocean University, Zhoushan 316000, China
Rockfish of the genus Sebastes, family Sebastidae, consist of approximately 110 species(Eitner et al., 1999; Love et al., 2002; Kai et al., 2003; Orr and Blackburn, 2004; Nelson, 2006) and are widelydistributed in cold waters throughout the north Pacific(Love et al., 2002; Kai et al., 2002a, b, 2003), northAtlantic(Nedreaas et al., 1994; Johansen et al., 2000, 2002; Roques et al., 2002), and across the southernhemisphere(Rocha-Olivares et al., 1999). SeveralSebastes species have been recorded in China(Liu and Qin, 1987; Jin, 2006), although S. koreanus(Kim and Lee, 1994)is only known from the southwestcoast of the Korean Peninsula and has not beenreported in China.
In the present study, Sebastes samples werecollected from the coastal waters of Dalian, Rongcheng, and Qingdao in China from May 2010 toSeptember 2011. The samples were morphologicallyidentified as S. hubbsi based on the findings ofprevious studies(Liu and Qin, 1987; Jin, 2006).However, the S. hubbsi description from China wasinconsistent with that from Japan and South Korea(Nakabo, 2000; Kim et al., 2005). The mitochondrialcytochrome oxidase subunit I(COI)gene noticeablyvaries between species but little within a given species(Gross, 2012). Therefore, a fragment of the COI gene, which is used for DNA barcoding(Hebert et al., 2003), has proven extremely effective fordiscriminating species(Domingues et al., 2013;Puckridge et al., 2013), discovering newly recorded and new species(Gao et al., 2011; Qin et al., 2013), uncovering cryptic species(Hajibabaei et al., 2007;Zemlak et al., 2009), and identifying species(Hebert et al., 2004; Bian et al., 2008; He et al., 2011).
By combining morphological and genetic analyses, the specimens in this study were identified as S. koreanus. Our objective was to describe S. koreanusbased on morphological characters and provide thecorrect DNA barcode. The results highlight the needfor caution when identifying species of the genusSebastes in China and will be helpful for fisheriesmanagement, biodiversity conservation, and sustainable exploitation of this species. 2 MATERIAL AND METHOD 2.1 Sampling
Samples were collected from the coastal waters ofDalian, Rongcheng and Qingdao in China duringMay 2010 to September 2011(Fig. 1). All individualswere identified based on morphological characteristics(Kim and Lee, 1994; Kim et al., 2005), and a piece ofmuscle tissue was obtained from each individual thatwas then preserved in 95% ethanol or by freezing.Additionally, tissues of 10 S. hubbsi individuals fromJapan were obtained(Fig. 1). All examined S. koreanusspecimens were frozen and preserved at the FisheryEcology Laboratory, Fisheries College, OceanUniversity of China in Qingdao.
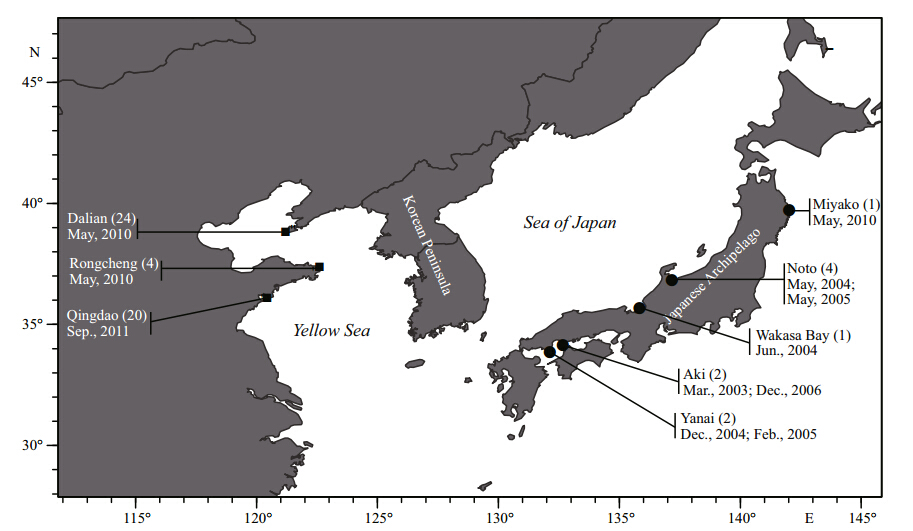 |
| Fig. 1 Sample collection information of Sebastes koreanus(■) and Sebastes hubbsi(●) |
Morphometric characters were compared withprevious S. koreanus and S. hubbsi records. Countsincluded: dorsal fin spines and rays, pectoral fin rays, anal fin spines and rays, pelvic fin spines and rays, lateral line scales, and vertebrae. Color and pigmentation were recorded by taking pictures offresh fish. 2.3 DNA extraction and sequencing
Genomic DNA was isolated from muscle tissue byproteinase K digestion followed by a st and ard phenolchloroformmethod. The fragment of mitochondrialDNA COI gene fragment was amplified using primersF1, 5′-TCAACCAACCACAAAGACATTGGCAC-3′, and R1, 5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′(Ward et al., 2005). All 10 of the S. hubbsi tissuesfrom Japan and 19 S. koreanus individuals, whichincluded 8 from Dalian, 4 from Rongcheng, and 7from Qingdao, were included. Polymerase chainreaction(PCR)was carried out in a 25-μL reactionmixture containing 17.5 μL of ultrapure water, 2.5 μLof 10×PCR buffer, 2 μL of dNTPs, 1 μL of each primer(5 μmol/L), 0.15 μL of Taq polymerase, and 1 μL of DNA template. PCR amplification was carriedout in a Biometra thermal cycler(Göttingen, Germany)under the following conditions: 5 mininitial denaturation at 95°C, and 30 cycles of 45 s at94°C for denaturation, 45 s at 52°C for annealing, and 45 s at 72°C for extension, and a final extension at72°C for 10 min. PCR products were purified with aGel Extraction Mini Kit(Watson BioTechnologiesInc., Shanghai, China). The purified product was usedas the template DNA for cycle sequencing reactionsperformed using a BigDye Terminator CycleSequencing Kit(ver. 2.0, PE Biosystems, Foster City, CA), and bi-direction sequencing was conducted onan ABI Prism 3730 automatic sequencer(AppliedBiosystems, Foster City, CA)using the same primersas those used for PCR amplification. 2.4 Sequence analysis
To determine the DNA barcode of S. koreanus, 33COI sequences were downloaded from GenBank foranalysis(Table 1). Sequences were aligned usingDNASTAR software(DNASTAR Inc., Madison, WI, USA). A neighbor-joining tree was constructed and distances between and within groups were determinedusing MEGA 5.0(Tamura et al., 2011)with 1 000bootstrap replicates based on genetic distances thatwere calculated using the best selected model K2P.
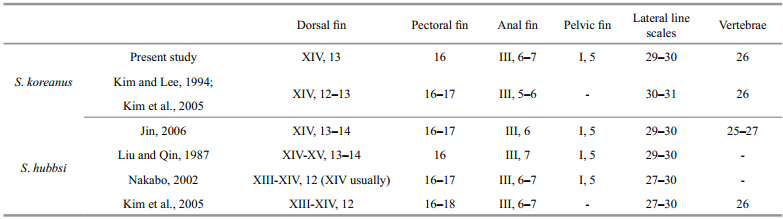
The general morphological features are shown inFig. 2. Body and head are moderately compressed.The fish are light brown with tiny dark spots on thebody, dorsal fin, pectoral fin, anal fin, pelvic fin, and caudal fin. Additionally, there are 4–5 wide indistinctvertical patterns on the side: 1 before the dorsal fin, 1at the caudal peduncle, and 3 in the middle of thebody. The fish have white spots on the base of dorsalfin. There are 2 radial stripes behind and below theeyes and 1 large dark blotch on the opercle. The 2ends of the upper chin reach the lower-middle of theeye. The fish have 14 dorsal fin spines, 13 dorsal finrays, 16 pectoral fin rays, 1 pelvic fin spine, 5 pelvicfin rays, 3 anal fin spines, 6–7 anal fin rays, 29–30lateral line scales, and 26 vertebrae. There are 5 spinescovering the preopercle, which is the bone that makesup the first part of the gill cover. Table 2 showssummaries of selected morphological characters of S. koreanus in the present and previous studies.
 |
| Fig. 2 Lateral views of Sebastes koreanus, 92.8 mm SL |
 |
A 625-bp COI gene fragment was obtained from29 individuals. All haplotype sequences weresubmitted to GenBank with the following accessionnumbers: KJ747353–KJ747356 for S. koreanus and KJ747357–KJ747358 for S. hubbsi. After combiningthe downloaded COI sequences of congeners, a totalof 62 sequences were used for the analysis. The meanK2P distance within S. koreanus was 0.3%, in contrastto the distance within the genus, which was 5.0%. Theminimum and maximum net genetic distancesbetween S. koreanus and other Sebastes species were 3.1% and 7.6%, respectively, both of which exceedthe threshold of species delimitation(~2%)(Hebert et al., 2003).
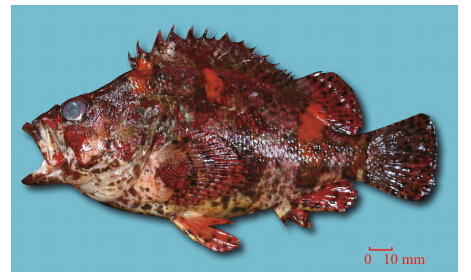 |
| Fig. 3 Lateral views of Sebastes hubbsi, 142 mm SL |
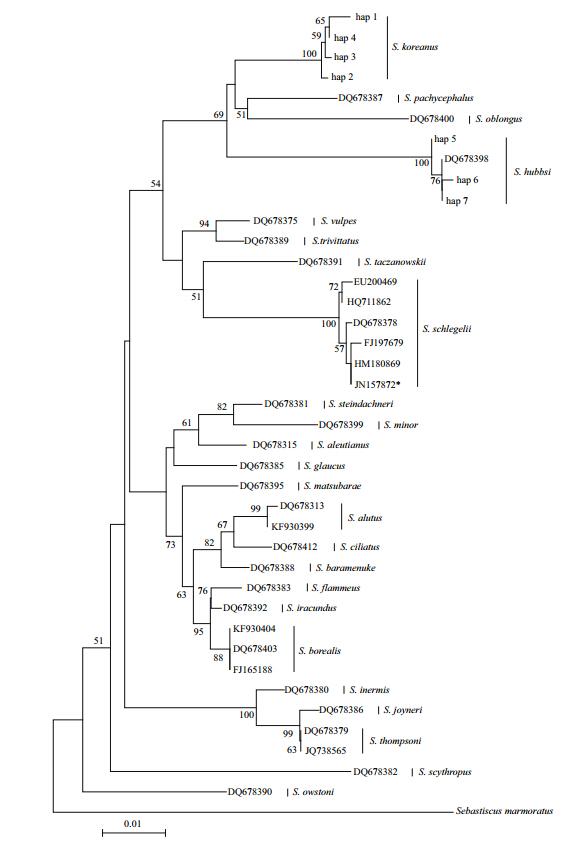
A neighbor-joining tree was constructed based onthe K2P model with 1 000 bootstrap replicates.Sebastiscus marmoratus was chosen as the out-groupto root the tree(Fig. 4). All the COI sequences ofSebastes koreanus clustered in the same group, and 4haplotypes were detected. One haplotype(Hap 1)wasunique to an individual from Qingdao. The secondhaplotype(Hap 2)was shared by 2 individuals fromQingdao. Four individuals from Dalian and 1individual from Qingdao shared the third haplotype(Hap 3). All of the remaining individuals shared thefourth haplotype(Hap 4). Additionally, all of the S .hubbsi COI sequences shared 3 haplotypes(Hap 5, Hap 6, and Hap 7) and clustered with the COIsequence that was downloaded from GenBank(DQ678398). The results indicate a large geneticdistance(4.8%)between S. koreanus and S. hubbsi .Additionally, the 5 Sebastes schlegelii COI sequencesclustered with the S. koreanus sequence(JN157872)from GenBank.
 |
|
Fig. 4 Neighbor-joining tree that was constructed using the K2P model for COI gene sequences of S. koreanus and 23 other Sebastes species Bootstrap values of >50% from 1 000 replicates are shown. *: S. koreanus COI sequence(JN157872)that was downloaded from GenBank. Sebastiscusmarmoratus was chosen as the out-group to root the tree. |
The morphological characters were photographed, counted, and then compared with previous records(Table 2). The present specimens were characterizedby 14 dorsal fin spines, 13 rays, 29–30 pored lateralline scales, 1 large dark blotch on the opercle, and 2radial stripes behind and below the eyes. Thesefindings are consistent with the descriptions of S .hubbsi from China(Liu and Qin, 1987; Jin, 2006) and S. koreanus from South Korea(Kim and Lee, 1994;Kim et al., 2005). Consequently, the specimens werefirst morphologically identified as S. hubbsi based onthe descriptions of previous reports(Liu and Qin, 1987; Jin, 2006). The description of S. hubbsi fromChina(Liu and Qin, 1987; Jin, 2006)is also differentfrom those in Japan and South Korea(Nakabo, 2000;Kim et al., 2005)but consistent with that of S. koreanus(Kim and Lee, 1994; Kim et al., 2005). Themost distinguishable character for these 2 species isbody color: S. koreanus is light brown with tinyscattered black spots, while S. hubbsi is a reddishbrown. Moreover, there are 2 radial stripes behind and below the eyes and 1 large dark blotch on theopercle for S. koreanus but not for S. hubbsi, and theshape of the anal, pelvic, and dorsal fins of S. koreanusare quite different from those of S. hubbsi(Figs.2, 3).Although the majority of morphometric charactersoverlap, the number of dorsal fin rays is a character that distinguishes S. koreanus(13)from S. hubbsi(12). Additionally, there are variations between S. koreanus in the present study and those recorded fromSouth Korea with regard to anal fin and lateral linescales. For the traits with overlapping ranges, it islikely that the specimens were in different life stages and this variation is natural. Based on these data, thespecimens in this study were morphologicallyidentified as S. koreanus.
The COI sequence is recognized as an effective and reliable method for identifying species(Hebert et al., 2003). It should be noted that the S. koreanus COIsequence from GenBank(JN157872)shared the samehaplotype with an S. schlegelii sequence(HM180869) and cluster within the S. schlegelii group; therefore, this specimen was most likely misidentified. Ouranalysis indicates that S. koreanus specimens in thisstudy can be differentiated from other Sebastesspecies because the net genetic distance ranged from3.1% to 7.6%. Moreover, there was a large geneticdistance(4.8%)between S. koreanus and S. hubbsi .These results provide strong support S. koreanus is avalid species at the genetic level. By combining themorphological and DNA sequences analyses, it canbe concluded that the S. hubbsi from previous studiesin China(Liu and Qin, 1987; Jin, 2006)are actually S. koreanus.
As shown in Fig. 1, this species is distributed in thenorthern coastal waters of China, which is consistentwith the indigenous distribution area for previouslydescribed S. hubbsi in China(Liu and Qin, 1987; Jin, 2006). It has previously been thought that S. koreanusis indigenous to the Korean Peninsula, which isinaccurate based on the results in this study.Additionally, studies on the fisheries biology(Wang et al., 2011) and karyotypes of S. hubbsi(Zheng et al., 1997), in which the sampling locations were HaizhouBay and Qingdao, respectively, were likely focusedon S. koreanus instead. Previous reports demonstratedthat S. koreanus were obtained from the Yellow Seaoff the southwest coast of South Korea(Kim and Lee, 1994; Yoon and Kim, 2007), which is almost at thesame latitude as Qingdao. Therefore, it can be inferredthat S. koreanus is distributed along the coastal watersof the southwest Korean Peninsula and northernChina through Liaoning Province and Sh and ongPeninsula. 5 CONCLUSION
A new record of S. koreanus from China isdocumented based on morphological characters and DNA barcoding. The morphological characters areconsistent with descriptions of this species fromSouth Korea. COI sequence analysis demonstratedthere are significant differences between S. koreanus and other species of the same genus, which shows thatS. koreanus is a valid species at the genetic level. S. koreanus was not thought to exist in China and wasrecorded as S. hubbsi(Liu and Qin, 1987; Jin, 2006).The purpose of the present study was to report a newrecord of S. koreanus from China as well as clarifyprevious species misidentification. Morphologically and ecologically, Sebastes species display greatdiversity in form and function. As a substantialamount of research has addressed the phylogeny and evolution of this genus(Nedreaas et al., 1994;Johansen et al., 2000, 2002; Roques et al., 2002; Kai et al., 2002a, 2002b, 2003, 2012; Hyde and Vetter, 2007; Yoon and Kim, 2007), accurate taxonomicdesignation is important. Consequently, we hope thatour results will aid in more explicit speciesidentification within the genus Sebastes in China and contribute to fisheries management, biodiversityconservation, and sustainable exploitation of S. koreanus . 6 ACKNOWLEDGEMENT
We deeply thank Dr. ZHANG H. for the samplecollection as well as Mrs. KANG S. H., Mr. LI Y., and Mr. QIN Y. for offering assistance with writing thepaper.
| Bian X D, Zhang X M, Gao T X, Xiao Y S. 2008. Morphological and genetic identification of Japanese halfbeak (Hyporhamphus sajori) eggs. J. Fish. Chin., 32 (3): 342-352. (in Chinese) |
| Domingues R R, de Amorim A F, Hilsdorf A W S. 2013.Genetic identification of Carcharhinus sharks from the southwest Atlantic Ocean (Chondrichthyes:Carcharhiniformes). J. Appl. Ichthyol., 29 (4): 738-742. |
| Eitner B, Kimbrell C, Vetter R. 1999. Sebastes moseri (Scorpaeniformes: Scorpaenidae): a new rockfish from the eastern North Pacific. Copeia, (1): 85-92. |
| Froese R, Pauly D. 2014. FishBase. World Wide Web electronic publication. http://www.fishbase.org. Accessed on 2014-1-4. |
| Gao T X, Ji D P, Xiao Y S, Xue T Q, Yanagimoto T, Setoguma T. 2011. Description and DNA barcoding of a new Sillago species, Sillago sinica (Perciformes: Sillaginidae), from coastal waters of China. Zool. Stud., 50 (2): 254-263. |
| Gross M. 2012. Barcoding biodiversity. Curr. Biol., 22 (3):R73-R76. |
| Hajibabaei M, Singer G A C, Hebert P D N, Hickey D A. 2007.DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends.Genet., 23 (4): 167-172. |
| He W P, Cheng F, Li Y X, Liu M, Li Z J, Xie S G. 2011.Molecular identification of Coilia ectenes and Coilia mystus and its application on larval species. Acta Hydr.Sin., 35 (4): 565-571. (in Chinese) |
| Hebert P D N, Ratnasingham S, de Waard J R. 2003. Barcoding animal life: cytochrome c oxidase subunit I divergences among closely related species. P. Roy. Soc. B., 270 (Suppl. 1): S96-S99. |
| Hebert P D N, Stoeckle M Y, Zemlak T S, Francis C M. 2004.Identification of birds through DNA barcodes. PLoS Biol., 2 (10): e312. |
| Jin X B. 2006. Fauna Sinica of China, Ostichthyes,Scorpaeniformes. Science Press, Beijing. p.154-156. (in Chinese) |
| Johansen T, Danıelsdóttir A K, Meland K, Nævdal G. 2000.Studies of the genetic relationship between deep-sea and oceanic Sebastes mentella in the Irminger Sea. Fish. Res., 49 (2): 179-192. |
| Johansen T, Daníelsdóttir A K, Nævdal G. 2002. Genetic variation of Sebastes viviparus Kroyer in the North Atlantic. J. Appl. Ichthyol., 18 (3): 177-180. |
| Hyde J R, Vetter R D. 2007. The origin, evolution, and diversification of rockfishes of the genus Sebastes (Cuvier). Mol. Phyl. Evol., 44 (2): 790-811. |
| Kai Y, Nakayama K, Nakabo T. 2002a. Genetic differences among three colour morphotypes of the black rockfish,Sebastes inermis, inferred from mtDNA and AFLP analyses. Mol. Ecol., 11 (12): 2 591-2 598. |
| Kai Y, Yagishita N, Ikeda H, Nakabo T. 2002b. Genetic differences between two color morphotypes of the redfish,Sebastes scythropus (Osteichthyes: Scorpaenidae).Species Divers., 7 (4): 371-381. |
| Kai Y, Nakayama K, Nakabo T. 2003. Molecular phylogenetic perspective on speciation in the genus Sebastes (Scorpaenidae) from the Northwest Pacific and the position of Sebastes within the subfamily Sebastinae.Ichthyol. Res., 50 (3): 239-244. |
| Kai Y, Park K D, Nakabo T. 2012. The incomplete history of mitochondrial lineages between two rockfishes, Sebastes longispinis and Sebastes hubbsi (Scorpaeniformes:Scorpaenidae). J. Fish. Biol., 81 (3): 954-965. |
| Kim I S, Choi Y, Lee C L, Lee Y J, Kim B J, Kim J H. 2005.Illustrated Book of Korean Fishes. Kyohak, Seoul. 221p. |
| Kim I S, Lee W O. 1994. A new species of the genus Sebastes (Pisces; Scorpaenidae) from the Yellow Sea, Korea.Korean J. Zool., 37 (3): 409-415. |
| Liu C X, Qin K J. 1987. Fauna Liaoningica Pisces. Liaoning Science and Technology Press, Shenyang. p.381-382. (in Chinese) |
| Love M S, Yoklavich M, Thorsteinson L. 2002. The rockfishes of the northeast Pacific. UC Press, Berkeley. 414p. |
| Nakabo T. 2000. Fishes of Japan with Pictorial Keys to the Species. Japanese, 2nd edn. Tokai Univ. Press, Tokyo,Japan. 594p. |
| Nedreaas K, Johansen T, Nævdal G. 1994. Genetic studies of redfish (Sebastes spp.) from Icelandic and Greenland waters. ICES J. Mar. Sci., 51 (4): 461-467. |
| Nelson J S. 2006. Fishes of the World. 4 th edn. Wiley, New Jersey. 624p. |
| Orr J W, Blackburn J E. 2004. The dusky rockfishes (Teleostei:Socrpaeniformes) of the North Pacific Ocean: resurrection of Sebastes variabilis (Pallas, 1814) and a redescription of Sebastes ciliatus (Tilesius, 1813). Fish. Bull., 102 (2): 328-348. |
| Puckridge M, Andreakis N, Appleyard S A, Ward R D. 2013.Cryptic diversity in flathead fishes (Scorpaeniformes:Platycephalidae) across the Indo-West Pacific uncovered by DNA barcoding. Mol. Ecol. Resour., 13 (1): 32-42. |
| Qin Y, Song N, Zou J W, Zhang Z H, Cheng G P, Gao T X,Zhang X M. 2013. A new record of a flathead fish (Teleostei: Platycephalidae) from China based on morphological characters and DNA barcoding. Chin. J. Oceanol. Limnol., 31 (3): 617-624. |
| Rocha-Olivares A, Rosenblatt R H, Vetter R D. 1999. Cryptic species of rockfishes (Sebastes : Scorpaenidae) in the Southern Hemisphere inferred from mitochondrial lineages. J. Hered., 90 (3): 404-411. |
| Roques S, Sévigny J M, Bernatchez L. 2002. Genetic structure of deep-water redfish, Sebastes mentella, populations across the North Atlantic. Mar. Biol., 140 (2): 297-307. |
| Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol.Evol., 28 (10): 2 731-2 739. |
| Wang Y J, Ye Z J, Zhang C, Dou S Z. 2011. The fisheries biology of Sebastes hubbsi in Che-Niushan Island.Periodic Ocean Univ. China, 41 (12): 46-52. (in Chinese) |
| Ward R D, Zemlak T S, Innes B H, Last P R, Hebert P D. 2005.DNA barcoding Australia's fish species. Phil. Trans. R.Soc. B., 360 (1462): 1 847-1 857. |
| Yoon J M, Kim J Y. 2007. Genetic differences and variation in black rockfish (Sebastes schlegelii) and hwanghae rockfish (S. koreanus) from the Yellow Sea. Genes Genom., 29 (4): 437-445. |
| Zemlak T S, Ward R D, Connell A D, Holmes B H, Hebert P D N. 2009. DNA barcoding reveals overlooked marine fishes. Mol. Ecol. Res., 9 (s1): 237-242. |
| Zheng J S, Wang M L, Zhu L Y, Dai J X. 1997. Studies on the karyotypes of Agrammus agrammus (Temminck et Schlegel) and Sebastes hu bbsi (Matsubara). Periodic Ocean Univ. China, 27 (3): 333-338. (in Chinese) |
 2015, Vol. 33
2015, Vol. 33



