Shanghai University
Article Information
- LI Chuanbiao (李传标) , DING Jun (丁俊) , ZHOU Lizhen (周立镇) , ZHANG Zhao (张昭) , Shengkang (李升康) , LIU Wenhua (刘文华) , WEN Xiaobo (温小波) _L
- Survey of cyanomyovirus abundance in Shantou coastal waters by g20
- Chinese Journal of Oceanology and Limnology, 2015, 33 (3) : 604-615
- http://dx.doi.org/10.1007/s00343-015-4168-9
Article History
- Received Jul. 14, 2014;
- accepted in principle Sep. 3, 2014;
- accepted for publication Oct. 14, 2014
2 Marine Biology Institute, Shantou University, Shantou 515063, China
Viruses are the most abundant biological component of marine microbial communities (Wilhelm and Suttle, 1999 ; Wommack and Colwell, 2000) , and approximately 10 23viral infections occur every second in the ocean (Suttle,2007) . What has been termed the “Third Age of Phage” (Mann,2005) can be viewed more broadly as a renewal of interest in environmental viruses,particularly in the viruses of microbes (Suttle,2005; Brussaard et al., 2008; Rohwer and Thurber, 2009; Danovaro et al., 2011) . Previous investigations have shown that viruses play key roles in the regulation of biological production,which shapes aquatic communities,determines ecosystem dynamics, and alters nutrient cycling and energy flow (Bergh et al., 1989; Suttle et al., 1990; Thingstad et al., 1993; Wilhelm and Suttle, 1999 ; Zwirglmaier et al., 2007) . Viruses also mediate gene transfer among microorganisms in natural aquatic ecosystems,predominantly through virus-mediated transduction and their shift between lytic and lysogenic cycles (Wilhelm and Suttle, 1999 ; Wommack and Colwell, 2000) .
Cyanophages infect many kinds of cyanobacteria,which are among the most abundant forms of marine picoplankton (Partensky et al., 1999) ,including Synechococcus,Prochlorococcus, and strains of other cyanobacterial genera (Sullivan et al., 2003) . Cyanophages contain a common region of DNA that encodes the capsid assembly protein g20, and they can reach levels of abundance of 10 3–10 5/mL in nearshore and off-shore waters,sometimes even reaching concentrations >10 6/mL (Matteson et al., 2011,2013) . Compelling evidence indicates that cyanophages help to prevent and control algal blooms,reduce environmental pollution, and play critical roles in maintaining the stability of marine ecosystems (Suttle et al., 1990) . Several studies of the whole metagenomics of samples from aquatic ecosystems have also shown that viruses contain large numbers of virulence genes encoding toxic peptides and cholera toxin,as well as genes involved in host invasion and metabolic and functional pathways (Waldor and Mekalanos, 1996; Bidle et al., 2007; Dinsdale et al., 2008) .
Morphological studies have shown that all the reported cyanophages can be classified into three viral families,Myoviridae,Siphoviridae, and Podoviridae,with most belonging to the family Myoviridae (Safferman et al., 1983; S and aa and Larsen, 2006) . Cyanomyoviruses are not only morphologically similar to the well-characterized coliphage T4,but also express g20 (Fuller et al., 1998) . Many μmolecular tools,including pulsed-field gel electrophoresis (Øvreas et al., 2003; Larsen et al., 2004; Sandaa et al., 2008) and total community DNA-DNA hybridization (Wichels et al., 1998) ,have been used to determine the extensive diversity of viruses. The analysis of signature genes based on clone libraries has also proved an effective way to determine viral diversity (Zhong et al., 2002) . Restriction fragment length polymorphism (RFLP) or denaturing gradient gel electrophoresis analyses have shown that cyanophages are genotypically diverse in their natural environments and that this diversity varies among different marine environments (Wang and Chen, 2004; Filee et al., 2005; Yan et al., 2010; Wang et al., 2011) . However,little has yet been reported on the cyanophage diversity in the coastal waters of Shantou,located in the northeast South China Sea.
In this study,samples were collected from four stations in the Shantou coastal waters, and the g20gene was amplified and used in a PCR-RFLP analysis to determine the diversity and phylogeny of the cyanomyoviruses in these samples. The abundances of cyanophage,heterotrophic bacteria, and picophytoplankton were assessed with flow cytometry and the virus-host contact rates were deduced from these data. Pearson’s correlation coefficients analyses indicated that synergistic and antagonistic relationships exist between the biotic and abiotic variables in the Shantou coastal waters. Our results provide a basis for further investigations of the virioplankton in the Shantou coastal waters. 2 MATERIAL AND METHOD 2.1 Sample collection
Water samples were collected from Niutianyang,Shantou Harbor,Nangang, and a freshwater reservoir at Shantou University in the period 16–21 June 2012 (Fig. 1) . We used an YSI 650 Sonde (YSI Instruments,Yellow Spring,OH) on a cruise to measure depth profiles,temperatures,pHs, and dissolved oxygen (DO) .
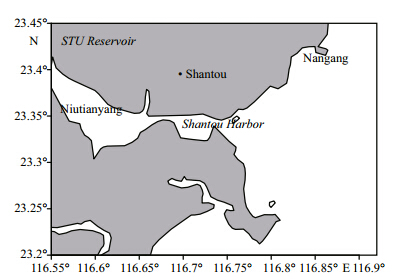 |
| Fig. 1 Location of the sampling stations in Shantou coastal waters
The four sampling sites (Niutianyang,Shantou Harbor,Nangang, and STU Reservoir) are labeled. |
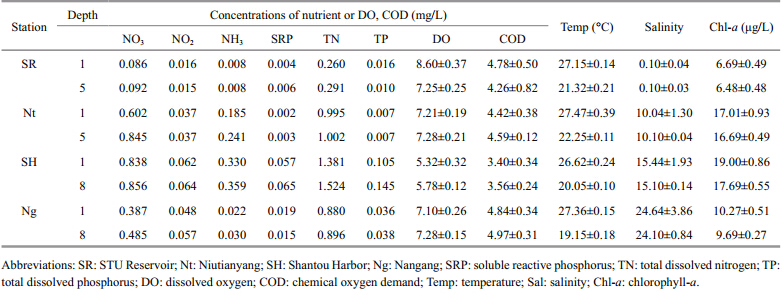
The concentrations of nutrients,including NO3-,NO2-,NH3,chemical oxygen dem and (COD) ,soluble reactive phosphorus (SRP) ,total dissolved nitrogen (TN) , and total dissolved phosphorus (TP) ,were measured in the water samples filtered on board through 0.45 m cellulose acetate membranes,which were then stored at -20°C until further analysis,according to the GB/T 12763.4-2007 (State Bureau of Quality and Technical Supervision,2007) .
Chlorophyll a (Chl- a) concentrations were determined in triplicate samples collected on polycarbonate filters with a 0.22 m pore size and 47 mm diameter. The samples were extracted in 90% acetone for 24 h at 4°C, and then centrifuged at 8 000 gfor 10 min. The absorbance spectra of the extracts were used to determine the Chl- aconcentrations (Jeffrey and Humphrey, 1975) . 2.2 Flow-cytometric analysis
The samples were analyzed with a FACSAria flow cytometer (Becton-Dickinson,Mountain View,CA) equipped with a blue laser beam fixed at 488 nm and the original filter setup. For the enumeration of picophytoplankton,5 mL of yellowish-green fluorescent beads of 1 μm diameter (fi nal dilution 10 -4; μmolecular Probes,San Francisco,CA) were used as the internal reference to calibrate cell fluorescence emissions and light scatter signals,allowing fluorescence and cell size to be compared among different samples. Each sample was run for 2 min at a rate of about 90 μL/min,with the discriminator set to red fluorescence (Marie et al., 1997) . To analyze the heterotrophic bacteria,the samples were diluted 50-fold with water from the same sampling site,filtered with 0.2 m pore filters, and stained with SYBR Green (Molecular Probes) at a final dilution of 10 -4of the commercial stock solution. The samples were incubated for 15 min in the dark, and then analyzed with the discriminator set to green fluorescence for 1 min at a rate of 50 mL/min. To analyze the viruses,dilutions (1:100) of the natural samples were added to TE buffer (10 mmol/L Tris,1 mmol/L EDTA,pH 7.8) . SYBR Green I was added to a final dilution of 10 -4, and the samples were incubated at 80°C in the dark for 15 min. Green fluorescence (fluorescein isothiocyanate) was used as the discriminator (Marie et al., 1999; Chen et al., 2001; Brussaard,2006; Wang et al., 2010) . The data were analyzed with the FCS Express V3 software (De Novo Software,Becton-Dickinson) . 2.3 PCR amplification of g20 fragments
The viral particles in the water samples were concentrated with a previously described protocol (Chen et al., 1996) . Briefly,40–80 L of each water samples was filtered through a Millipore filter (pore size,0.45 m) in a ProFlux M-12 system (Millipore,Boston,USA) . The virus concentrates were stored in the dark at 4°C. The viral pellets were resuspended in 200 L of distilled water and frozen at -20°C. The sample DNA was extracted with phenol-chloroform extraction,followed by ethanol precipitation. Two sets of primers,CPS1/CPS4 and CPS1/CPS8,were used to amplify overlapping regions of the g20 gene from cyanophages,with expected fragment lengths of about 470 bp and 592 bp (Fuller et al., 1998; Marston and Sallee, 2003) . The 25 L reaction mix contained 10 Taqreaction buffer,1.5 mmol/L MgCl 2,250 μmol/L each deoxynucleoside triphosphate,10 μmol/L each primer,1.25 U of Taq DNA polymerase (TaKaRa,Dalian,China) , and 1 L of template DNA. The PCR cycling conditions consisted of an initial denaturation step at 94°C for 5 min,followed by 35 cycles of 94°C for 45 s,47°C for 45 s, and 72°C for 45 s, and a final extension at 72°C for 5 min. The PCR products were analyzed by electrophoresis on a 1.0% agarose gel. 2.4 Library construction and RFLP analysis
The PCR products were ligated into the pMD19-T vector and the resulting reactions were used to transform Escherichia coli DH5 cells (TaKaRa) ,according to the manufacturer’s instructions. The cyanophage genotypes were distinguished using a PCR-RFLP technique. The PCR products amplified with primer set CPS1/CPS8 or CPS1/CPS4 were digested with a restriction enzyme (RsaI,MspI,or HinfI) (Invitrogen,Carlsbad,CA) and the digestion products were separated by electrophoresis on a 3.0% agarose gel. Clones containing g20gene fragments with different PCR-RFLP profiles were picked and their genes sequenced.
A BLAST analysis of the nucleotide sequences of g20was performed through the National Center of Biotechnology Information (NCBI) website. Regions of the sequences with the strongest identity were chosen for phylogenetic analysis. The sequences were aligned with ClustalW (Thompson et al., 1997) and a phylogenetic tree was constructed with the neighborjoining (NJ) method using the MEGA 4.1 software package (Arizona State University,AZ,USA) . Coliphage T4 and other reference sequences were used as outgroups. A phylogenetic tree constructed with the maximum parsimony approach in MEGA 4.1 yielded similar results. The node reproducibility in the tree topology was estimated with a bootstrap analysis that included 100 replicate data sets. 2.5 Quantitative PCR of g20
The samples for qPCR analysis were collected in 5 mL cryovials,immediately frozen in liquid nitrogen, and stored at -80°C until analysis. The primer set CPS1/CPS4 was used to estimate the g20gene copy numbers. Synechococcusphage P77 (g20 accession AY027982) DNA was purified and used to construct a linear st and ard curve,according to Matteson et al. (2011) . Each 25 L quantitative PCR (qPCR) mixture contained 12.5 L of SYBR Green I real-time PCR Master Mix (TaKaRa) ,0.8 L of the CPS1 forward primer (10 μmol) ,0.8 L of the CPS4 reverse primer (10 μmol) , and 2 L of viral sample or st and ard. Realtime PCR was performed in a Lightcycler 480 (Roche,Alameda,CA) with the program: 95°C for 30 s,followed by 40 cycles of 95°C for 15 s and 60°C for 31 s. 2.6 Virus-host contact rates
The contact rates for the total viral community and for the cyanomyovirus population with the heterotrophic bacteria and cyanobacteria,including Synechococcus and Prochlorococcus,were calculated according to Murray and Jackson (1992) : contacts= (2Sπ ωDv) VB,where VB is the virus-tobacteria ratio. The average diameter (ω) of heterotrophic bacteria (0.45 10 -4cm) was used (Lee and Fuhrman, 1987) , and a diameter of 1.0 10 -4cm for Synechococcus and 0.5 10 -4cm for Prochlorococcus (Waterbury et al., 1986) . The diffusivity of the virus particles (Dv) was assumed to be 3.45610 -3cm2/d (Murray and Jackson, 1992) and a Sherwood number (S) of 1.06 was used (Wilhelm et al., 1998) . The specific contact rates of viruses per bacterium per day were determined by dividing the contact rate by the host abundance. 2.7 Statistical analysis
Percentage recoveries from spiked samples were analyzed with the equation: percentage recovery= (measured copies in water sample spiked with phage genomic DNA-average measured copies in unspiked water samples) / (viral genomic copies added) 100%. The relationships between the biological parameters and the g20gene copy number were compared using regression and correlation data on samples that were untransformed or logtransformed using Pearson’s pairwise statistics in the NCSS Statistical Software (Kaysville,UT) . The data for relative mRNA expression were analyzed with the 2-ΔΔCtmethod (Livak and Schmittgen, 2001) . All data are expressed as means±SE. The data were analyzed with one-way ANOVA, and P<0.05 were considered statistically significant. 2.8 Data deposition
The DNA sequences acquired in this study were deposited at the NCBI GenBank website under accession numbers HG326431–HG326491. 3 RESULT 3.1 Water conditions
Water samples from the upper (0–1 m depth) and lower water layers (5–8 m depth) were collected in the period 16–21 June 2012 at four stations (Niutianyang,Shantou Harbor,Nangang, and STU Reservoir) in the coastal waters of Shantou,northeast South China Sea (Fig. 1) . Salinity varied dramatically,with a range of 0.1–24.64 (0.1 in the STU Reservoir and 24.64 at the Nangang station) . The summer temperatures were warm in the upper layers (26.7–27.5°C) and cool in the lower layers (19.15–22.25°C) . The nutrient concentrations were highest at Niutianyang station. NO3-was the most significant nitrogen source (relative to NO 2 and NH4+) at the four stations. There were relatively high concentrations of SRP,TN, and TP at the Shantou Harbor station,whereas the concentrations of DO and COD were low. The biomass of primary producers was highest at the STU Reservoir (total Chl- aconcentrations: up to 19.00 μg/L at 1 m depth and 17.69 μg/L at 5 m depth) and lowest in Shantou Harbor (Chl- a concentrations: 6.69 μg/L at 1 m depth and 6.48 μg/L at 8 m depth) . Overall,the hydrological parameters did not differ significantly between the upper and lower layers at the four sample stations (Table 1) ,so we mixed the upper and the lower layers at each of the four sampling stations in the subsequent analyses.
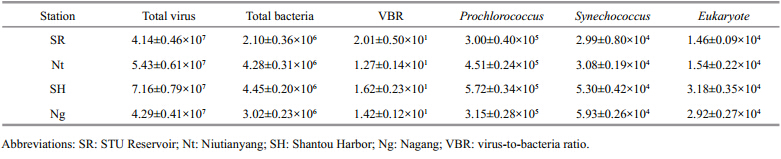
The abundances of viruses,heterotrophic bacteria, and picophytoplankton in the Shantou coastal waters were monitored in the summer of 2012 (Table 2) . Although we observed shifts in the phytoplankton and bacterial biomasses,the abundance of viruses remained relatively constant at each of the four stations examined. The concentration of the total virus particles ranged from 4.14 10 7to 7.16 10 7/mL. Phytoplankton (Prochlorococcus,Synechococcus,eukaryotes) were most abundant in Shantou Harbor,with concentrations of up to 5.72 10 5/mL,5.30 10 4/mL, and 3.18 10 4/mL,respectively. In contrast,STU Reservoir had the lowest concentrations of phytoplankton,at 3.00 10 5/mL,2.99 10 4/mL, and 1.46 10 4/mL,respectively. In the summer,the virusto-bacterium ratio (VBR) ranged from 12.7 to 20.1,with the highest ratio observed at the STU Reservoir and the lowest at the Niutianyang station.
In total,126 clones were amplified and identified as the g20fragment. An RFLP analysis and subsequent sequencing was used to identify the unique genotypes of these g20fragments. Ultimately,61 unique sequences were obtained after a filtering procedure was applied to remove extremely similar sequences,with a criterion cutoff of 97%. In this study,6,19,16, and 20 genotypes were retrieved from the clone libraries constructed from the samples from the STU Reservoir,Niutianyang,Shantou Harbor, and Nangang,respectively. A BLASTn analysis indicated that the nucleotide sequence similarities of the cyanomyovirus g20sequences extracted from the samples from Shantou coastal waters ranged from 47.0% to 97%.
A phylogenetic analysis of the cyanomyovirus g20sequences using both NJ and maximum parsimony methods revealed that the g20clones could be grouped into at least eight distinct clusters (Clusters I–III,A,C,E,N4, and W,containing 15,5,6,2,1,1,22, and 9 sequences,respectively) ,which corresponded to clones and phage isolates reported previously (Fig. 2) .
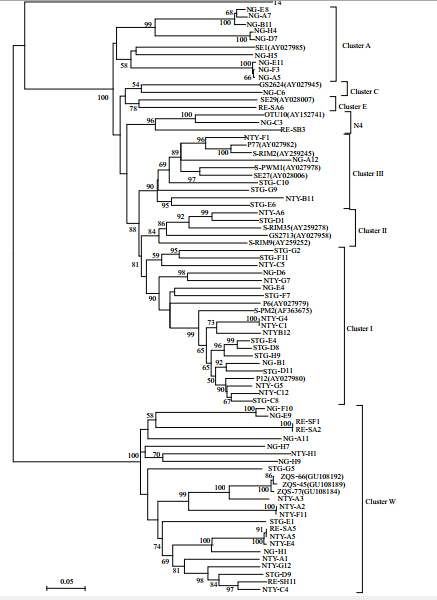 |
| Fig. 2 Neighbor-joining tree showing the phylogenetic relationships of cyanophages from Shantou coastal waters and other viruses isolated in previous studies
The tree was constructed on the basis of aligned cyanophage nucleotide sequences of the g20gene,with the T4 phage sequence used as the outgroup. “Clusters” refers to the classification scheme of Zhong et al. (2002) . The numbers in parentheses are the accession numbers of the nucleotide sequences. Boldface indicates cyanophage sequences from Shantou coastal waters. Bootstrap values <50 are not shown. The scale bar represents the number of nucleotide substitutions per residue. |
In the clone library from Niutianyang station,nine,seven, and two clones were affiliated with Cluster W,Cluster I, and Cluster III,respectively,whereas in the clone library from Shantou Harbor,three,eight, and three clones were affiliated with Cluster W,Cluster I, and Cluster III,respectively. In the clone library from Nangang,six,three, and one clones were affiliated with Cluster W,Cluster I, and Cluster III,respectively, and nine clones belonged to Cluster A. Thus,the Cluster W and Cluster I genotypes predominated in the viral communities in Shantou coastal waters. These data indicate that the diversity and the dominant components in the viral community of the Shantou coastal waters varied significantly. 3.4 g20gene copy numbers
Putative cyanomyovirus abundance,estimated by the quantitative PCR measurement of g20 gene copies,varied from 8.00 10 5/mL in the STU Reservoir to 5.65 10 6/mL at the Niutianyang station,whereas at the other two stations,Shantou Harbor and Nangang,the estimated cyanomyovirus abundance was 2.99 10 6/mL and 2.10 10 6/mL,respectively (Fig. 3) . Therefore,the abundance of the putative cyanomyoviruses at Niutianyang station was as much as seven-,three-, and two-fold higher than that in the STU Reservoir,Nangang, and Shantou Harbor,respectively (P<0.05) . However,there was nosignificant difference in the cyanomyoviral abundance between Shantou Harbor and Nangang (P>0.05) . Thus,putative cyanomyoviruses accounted for 1.92% (STU Reservoir) to 10.40% (Niutianyang) of the total viral populations in the summer samples from Shantou coastal waters.
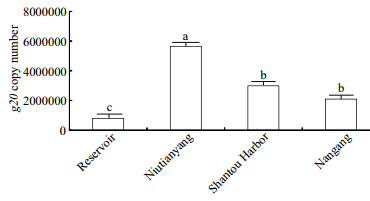 |
| Fig. 3 g20copy numbers at the four sampling sites Data (means±SD,n=3) at the different sampling sites with different letters are significantly different (P<0.05) . |
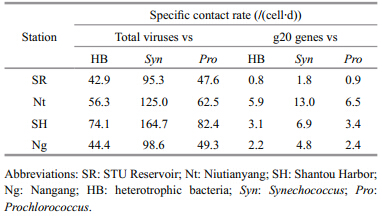
Overall,the contact rates between viruses and bacteria in the Shantou coastal waters varied dramatically,depending on the sampling location. We calculated the cell-specific contact rates between the host (heterotrophic bacteria,Synechococcus, and Prochlorococcus) and virus (total viral particles,putative cyanomyoviruses) (Table 3) . The rates ranged from 42.87 to 74.13 contacts/ (cell∙d) (average 54.40) between the heterotrophic bacterial communities and the total viruses in summer in the Shantou coastal waters. The rates for Synechococcusranged from 95.27 to 164.74 contacts/ (cell∙d) (average 120.90) ,whereas the rates for Prochlorococcusranged from 47.64 to 82.37 contacts/ (cell∙d) (average 60.45) .
In contrast,the cell-specific contact rates between putative cyanomyoviruses and the total bacteria ranged from 0.83 to 5.85 contacts/ (cell∙d) (average 2.98) . For Synechococcus and Prochlorococcus,the rates ranged from 1.83–13.00 contacts/ (cell∙d) (average 6.63) and 0.91–6.50 contacts/ (cell∙d) (average 3.31) ,respectively.
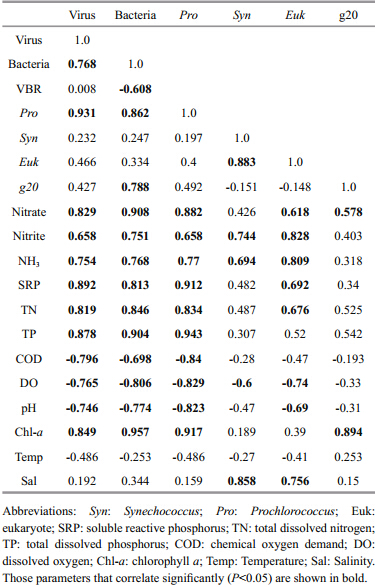
The virioplankton abundance correlated significantly positively with bacterial abundance,Prochlorococcusabundance,nitrate,TP,TN,SRP, and Chl- acontent,but negatively with COD,DO, and pH. Bacterial abundance and Prochlorococcusabundance correlated significantly positively with NO3-,NO2-,TP,TN,SRP, and Chl- acontent,but negatively with COD,DO, and pH. Synechococcusabundance correlated significantly positively with NO2-,NH3, and salinity,but negatively with DO. Eukaryote abundance correlated significantly positively with several nutrient factors (including NO3-,NO2-,NH3,AP,TN, and TP) and salinity,but negatively with DO and pH. Cyanomyovirus abundance correlated significantly positively with bacterial abundance,NO3-, and Chl- acontent (Table 4) .
 |
The distribution of viruses in marine ecosystems and their potential roles in biogeochemical cycles have been well documented in the last two decades (Suttle et al., 1990) . Increasing evidence has shown that marine viral activity is a driving force in maintaining the genetic diversity of bacterial communities and profoundly influences the functions of marine ecosystems (Weinbauer and Rassoulzadegan, 2004) . Currently,cyanophages from all habitats are classified into three viral families, Myoviridae,Siphoviridae, and Podoviridae,according to their morphologies and structures (Suttle et al., 1990) . Previous studies have demonstrated that Synechococcus and ,to a lesser extent,low-light adapted Prochlorococcusspp. are infected by cyanomyoviruses more often than by cyanopodoviruses or cyanosiphoviruses (Lu and Hodson, 2001; Sullivan et al., 2003) . In this study,the cyanomyovirus concentrations,inferred from g20gene copy numbers,showed that they are abundant and represent a substantial proportion of the total viral community in the coastal waters of Shantou. Surprisingly high densities of cyanomyoviruses (from 8.00 10 5/mL in the STU Reservoir to 5.65 10 6/mL at Niutianyang station) were found in these waters. High densities of cyanomyoviruses (2.4±1.5 10 4/mL,n=16 stations) have also been found in the upper mixed layer of the Sargasso Sea (Matteson et al., 2013) . A metagenomic survey of seawater from the Sargasso Sea revealed abundant cyanophage genes related to cyanomyoviruses (Venter et al., 2004) . In another study based on the most probable number assay,the authors demonstrated 10 3–10 4infectious particles in the coastal and pelagic waters of the Sargasso Sea (Waterbury et al., 1986) . Similarly,an average cyanomyovirus abundance of 3.2±2.7 10 5/mL (n=16 stations) was observed in the southern Pacific Ocean (Matteson et al., 2013) . Based on the data from our study,the abundance of cyanomyoviruses in the coastal waters of Shantou was clearly much higher than those in the Sargasso Sea,the coastal and pelagic waters of the Sargasso Sea, and the southern Pacific Ocean. This difference could be explained by the higher primary productivity and the more abundant heterotrophic bacteria in the Shantou inshore waters. Moreover,the percentages of cyanomyoviruses within the viral communities in the coastal waters of Shantou were particularly high: 1.92% in the STU Reservoir and 10.40% at the Niutianyang station. High proportions of cyanomyoviruses were also observed in the Sargasso Sea and the southern Pacific Ocean (Matteson et al., 2013) .
The variability of cyanophage communities is believed to correlate with differences in their host communities,based on the possible “kill-the-winner” approach,where the most abundant host population is reduced by its viral predators (Thingstad and Lignell, 1997) . In this study,although the cyanobacterial communities in the coastal waters of Shantou were not examined,several reports have indicated that the cyanobacterial communities in aquatic environments vary spatially (Palenik,1994; Kim and Lee, 2006) . The dynamic changes in cyanophage communities may indicate that their hosts,the cyanobacterial communities,also fluctuate in the coastal waters of Shantou.
In this study,we demonstrated that the cyanomyoviral communities in the coastal waters of Shantou are distinctly different from those reported in other aquatic and marine environments (Fuller et al., 1998; Wilhelm et al., 2006; Wang et al., 2010) (Fig. 2) . A phylogenetic analysis showed that their g20sequences were very diverse and could be distributed into eight distinct operational taxonomic units (OTUs) ,designated Clusters I–III,A,C,E,N4, and W. About 24.5%,8.2%, and 9.8% of the g20 clones isolated in this study belong to Clusters I,II, and III,respectively (Wang et al., 2010) ,whereas approximately 36.1%,14.7%, and 3.3% of the clones belonged to Clusters N4,W, and A,respectively (Fig. 2) . Most cyanophage communities from paddy floodwaters have been reported to forms eight unique groups (PFW-I to PFW-VI,CSP-PFW1, and CSPPFW2) (Wang et al., 2010) ,which are distinctly different from those in the paddy field soils (Wang et al., 2011) . The authors speculated that the differences in these two cyanophage communities suggest that their hosts,the cyanobacterial communities,also differ between soil and water types. The results of our study suggest that the cyanophage communities in Shantou coastal waters differ from those in aquatic and other marine environments. Overall,the phylogenetic structure of the g20sequences in the coastal waters of Shantou was distinctly different from that found in the South China Sea (our unpublished data) . Similarly,the g20sequences in an estuary (Zhong et al., 2002) and in Chesapeake Bay (Wang and Chen, 2004) differed from those in the open ocean.
It is surprising that a substantial proportion of the g20clones from the coastal waters of Shantou belonged to Cluster W. Members of Cluster W were also found in the coastal waters of Qingdao (Yan et al., 2010) . We hypothesize that the presence of Cluster W in the coastal waters of Shantou can be attributed to either slight pollution of the seawater or the appearance of novel Synechococcusstrains that are resistant to eutrophicated water (Zhong et al., 2002; Yan et al., 2010) .
The amplification of the g20gene products with primers CPS1/CPS8 from samples collected deep in the cyanobacterium-sparse Chukchi Sea suggested that this primer set targets bacteriophages other than those infecting cyanobacteria (Short and Suttle, 2005) . The g20genes amplified with primers CPS1/CPS8 are estimated to be of cyanophage origin because there is no direct evidence that the sequences of isolated noncyanophages can be amplified with these primers (Dorigo et al., 2004; Wilhelm et al., 2006; Wang et al., 2010) . Because the clones isolated in this study clustered with cyanophage clones from freshwater and seawater (Fig. 2) ,their hosts were presumed to be cyanobacteria. However,not all the PCR products amplified with the primer sets CPS1/CPS8 and CPS1/CPS4 were g20genes. In our study,126 clones with different nucleotide sequences were identified as g20fragments, and after the highly identical sequences were removed with a criterion cutoff of 97%,61 of them were new g20 sequences. The cyanophage communities in the coastal waters of Shantou were not constant across the four sampling sites and also differed from the cyanophage communities detected in the coastal waters of other regions (Yan et al., 2010) .
The contact rate provides an upper limit to the number of cells that can be infected by viruses (Suttle et al., 1990) . The viruses and closely related hosts are distributed widely across environments, and horizontal gene exchange occurs among phage communities from very different environments (Short and Suttle, 2005) . The contact rates between the total virus community and the heterotrophic bacteria were compared, and the rate of 42.87–74.13 contacts/ (cell∙d) (average 54.40) in the Shantou coastal waters in summer is similar to the total-virus–bacteria contact rates observed in the Pacific Ocean (21–164/ (cell∙d) ; Wang et al., 2011) and in eutrophic Masan Bay in Korea (12–321/ (cell∙d) ; Choi et al., 2003) ,but much higher than those observed in the Sargasso Sea (0.21–2.01 contacts/ (cell∙d) ; Wang et al., 2011) and the Gulf of Mexico (0.20–4.0/ (cell∙d) ; Wilhelm et al., 2003) . Prochlorococcusis distributed in the high-latitude South China Sea along the Kuroshio Current,the second largest warm-water current in the world (Jiao and Yanhui, 2002) . The contact rates between Prochlorococcuscells and cyanomyoviruses or the total virus community were nearly equal to these between cyanomyoviruses and heterotrophic bacteria at all four stations in the coastal waters of Shantou. In contrast,the contact rates between Synechococcuscells and cyanomyoviruses or the total virus community were much greater than those between cyanomyoviruses and heterotrophic bacteria in this region (Table 3) . This is partly attributable to the larger estimated diameter of this cyanobacterium (Matteson et al., 2013) . The high abundance of Synechococcus and Prochlorococcusat our sampling stations also indicates that cyanophage isolates infect picoplankton in this region.
Pearson’s correlation coefficients for the pooled parameters in the coastal waters of Shantou identified significant positive and negative relationships between the biotic and abiotic variables (Table 4) . Notably,the abundance of cyanomyovirus correlated significantly with the abundance of heterotrophic bacteria,nitrate, and the Chl- aconcentration in the coastal waters. These correlations between biotic and abiotic variables suggest the presence of both synergistic and antagonistic effects among microbial groups (Wang et al., 2010) .
Among the phytoplanktonic prokaryotes,members of the cyanobacteria (including Synechococcus and Prochlorococcus) are abundant,diverse, and widespread worldwide in different kinds of aquatic environments, and are potential hosts for cyanomyoviral infection (Sullivan et al., 2003) . Cooccurring cyanophages can modulate cyanobacterial communities and cyanophage infection can exert a major influence on cyanobacterial succession (Zhang et al., 2013) . Several previous studies have reported covariation between Synechococcus and cyanomyovirus abundances (Wang et al., 2011) ,indicating a tight coupling between the two communities. However,in most cases,cyanomyovirus abundance does not correlate with the abundance of cyanobacteria,suggesting a weak relationship and low viral impact on the temporal dynamics of cyanobacterial abundance. This weak viral impact on cyanobacteria was also seen in our study.
This was a preliminary study to investigate the cyanomyoviruses and other picoplankton in the Shantou coastal waters. Investigation of the spatial and temporal changes in the cyanomyoviral communities in these coastal waters,using more sampling sites and in different seasons (mainly in summer and winter) ,is in progress by our group. To the best of our knowledge,this is the first report of the total viral communities and cyanomyovirus abundance in the coastal waters of Shantou. The phylogenetic diversity revealed by the analysis of the viral capsid assembly protein gene g20shows that the cyanophages are diverse and distinctly different in freshwater lakes and marine environments. Our results suggest that cyanomyoviruses are ubiquitous and are an abundant component of the virioplankton in the Shantou coastal waters. 5 CONCLUSION
In this study,we have presented quantitative information about the cyanomyoviral communities in the coastal waters of Shantou. A phylogenetic analysis showed that the cyanomyoviral g20sequences in Shantou coastal waters were very diverse and could be assigned to eight distinct Clusters,which were different from those in freshwater lakes and marine environments. To the best of our knowledge,this is the first report of the total viral communities and cyanomyoviral abundance in the coastal waters of Shantou. This information about cyanomyoviral abundance and activities will extend our underst and ing of the microbial ecology in the region. 6 ACKNOWLEDGMENT
We thank Prof. WEI Chiju of Shantou University for his critical discussion and proof-reading of the manuscript. The authors thank Dr. LI Ping and Mr. HUANG Zhongwen for their assistance with the collection of samples.
| "Bergh Ø, BØrsheim K Y, Bratbak G, Heldal M. 1989. High abundance of viruses found in aquatic environments.Nature, 340 (6233) : 467-468. |
| Brussaard C P D, Wilhelm S W, Thingstad F, Weinbauer M G,Bratbak G, Heldal M, Kimmance S A, Middelboe M,Nagasaki K, Paul J H, Schroeder D C, Suttle C A, Vaqué D, Wommack K E. 2008. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME. J., 2 : 575-578. |
| Brussaard C P D. 2006. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ.Microbiol., 70 (3) : 1 506-1 513. |
| Bidle K D, Haramaty L, Barcelos E R J, Falkowski P. 2007.Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc.Natl. Acad. Sci. USA, 104 (14) : 6 049-6 054. |
| Chen F, Lu J R, Binder B J, Liu Y C, Hodson R E. 2001. |
| Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR gold.Appl. Environ. Microbiol., 67 (2) : 539-545. |
| Chen F, Suttle C A, Short S M. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ.Microbiol., 62 (8) : 2 869-2 874. |
| Choi D H, Hawang C Y, Cho B C. 2003. Comparison of virusand bacterivory-induced bacterial mortality in the eutrophic Masan Bay, Korea. Aquat. Microb. Ecol., 30 : 117-125. |
| Danovaro R, Corinaldesi C, Dell'anno A, Fuhrman J A,Middelburg J J, Noble R T, Suttle C A. 2011. Marine viruses and global climate change. FEMS Microbiol. Rev., 35 (6) : 993-1 034. |
| Dinsdale E A, Edwards R A, Hall D, Angly F, Breitbart M, Brulc J M, Furlan M, Desnues C, Haynes M, Li L L, McDaniel L, Moran M A, Nelson K E, Nilsson C, Olson R, Paul J, Brito B R, Ruan Y J, Swan B K, Stevens R,Valentine D L, Thurber R V, Wegley L, White B A,Rohwer F. 2008. Functional metagenomic profiling of nine biomes. Nature, 452 (7187) : 629-632. |
| Dorigo U, Jacquet S, Humbert J F. 2004. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl. Environ. Microbiol., 70 (2) : 1 017-1 022. |
| Filee J, Tetart F, Suttle C A, Krisch H M. 2005. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc. Natl. Acad. Sci. USA, 102 (35) : 12 471-12 476. |
| Fuller N J, Wilson W H, Joint I R, Mann N H. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g 20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol., 64 (6) : 2 051-2 060. |
| Jeffrey S W, Humphrey G F. 1975. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem.Physiol. Pflanz., 167 : 191-194. |
| Jiao N Z, Yang Y H. 2002. Ecological studies on Prochlorococcus in China seas. Chin. Sci. Bull., 47 (15) : 1 243-1 250. |
| Kim J D, Lee C G. 2006. Diversity of heterocystous filamentous cyanobacteria (blue-green algae) from rice paddy fields and their differential susceptibility to ten fungicides used in Korea. J. Microbiol. Biotech., 16 (2) : 240-246. |
| Larsen A, Flaten G A F, Sandaa R A, Castberg T, Thyrhaug R,Erga S R, Jacquet S, Bratbak G. 2004. Spring phytoplankton bloom dynamics in Norwegian coastal waters: microbial community succession and diversity.Limnol. Oceano., 49 (1) : 180-190. |
| Lee S, Fuhrman J A. 1987. Relationships between biovolume and biomass of naturally derived marine bacterioplankton.Appl. Environ. Microbiol., 53 (6) : 1 298-1 303. |
| Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCT method. Methods, 25 (4) : 402-408. |
| Lu J R, Chen F, Hodson R E. 2001. Distribution, isolation, host specificity, and diversity of cyanophages infecting marine Synechococcus spp. in river estuaries. Appl. Environ.Microbiol., 67 (7) : 3 285-3 290. |
| Mann N H. 2005. The third age of phage. Plos Biol., 3 (5) : e182. |
| Marie D, Partensky F, Jacquet S, Vaulot D. 1997. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol., 63 (1) : 186-193. |
| Marie D, Brussaard C P D, Thyrhaug R, Bratbak G, Vaulot D. 1999. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ.Microbiol., 65 (1) : 45-52. |
| Marston M F, Sallee J L. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters.Appl. Environ. Microbiol., 69 (8) : 4 639-4 647. |
| Matteson A R, Loar S N, Bourbonniere R A, Wilhelm S W. 2011. Molecular enumeration of an ecologically important cyanophage in a Laurentian Great Lake. Appl. Environ.Microbiol., 77 (19) : 6 772-6 779. |
| Matteson A R, Rowe J M, Ponsero A J, Pimentel T M, Boyd P W, Wilhelm S W. 2013. High abundances of cyanomyoviruses in marine ecosystems demonstrate ecological relevance. FEMS Microbiol. Ecol., 84 (2) : 223-234. |
| Murray A G, Jackson G A. 1992. Viral dynamics: a model of the effects of size shape, motion and abundance of singlecelled planktonic organisms and other particles. Mar.Ecol. Prog. Ser., 89 : 103-116. |
| Øvreas L, Bourne D, Sandaa R A, Casamayor E O, Benlloch S,Goddard V, Smerdon G, Heldal M, Thingstad T F. 2003. |
| Response of bacterial and viral communities to nutrient manipulations in seawater mesocosms. Aquat. Microb.Ecol., 31 : 109-121. |
| Palenik B. 1994. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl.Environ. Microbiol., 60 (9) : 3 212-3 219. |
| Partensky F, Hess W R, Vaulot D. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance.Microbiol. Mol. Biol. Rev., 63 : 106-127. |
| Rohwer F, Thurber R V. 2009.Viruses manipulate the marine environment. Nature, 459 (7244) : 207-212.. |
| Safferman R S, Cannon R E, Desjardins P R, Gromov B V,Haselkorn R, Sherman L A, Shilo M. 1983. Classification and nomenclature of viruses of cyanobacteria.Intervirology, 19 (2) : 61-66. |
| Sandaa R A, Larsen A. 2006. Seasonal variations in virus-host populations in Norwegian coastal waters: Focusing on the cyanophage community infecting marine Synechococcus spp. Appl. Environ. Microbiol., 72 (7) : 4 610-4 618. |
| Sandaa R A, Clokie M, Mann N H. 2008. Photosynthetic genes in viral populations with a large genomic size range from Norwegian coastal waters. FEMS Microbiol. Ecol., 63 (1) : 2-11. |
| Short C M, Suttle C A. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ.Microbiol., 71 (1) : 480-486. |
| Sullivan M B, Waterbury J B, Chisholm S W. 2003.Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature, 424 (6952) : 1 047-1 051. |
| State Bureau of Quality and Technical Supervision. 2007. The Specification for Oceanographic Survey-Observations of Chemical Parameters in Sea Water. China Standard Press,Beijing, China. (in Chinese) Suttle C A. 2005. Viruses in the sea. Nature, 437 (7057) : 356-361. |
| Suttle C A. 2005. Viruses in the sea. Nature, 437 (7057) : 356-361. |
| Suttle C A. 2007. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol., 5 (10) : 801-812. |
| Suttle C A, Chan A M, Cottrell M T. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature, 347 (6292) : 467-469. |
| Thingstad T F, Heldal M, Bratbak G, Dundas I. 1993. Are viruses important partners in pelagic feed webs?. Trends Ecol. Evol., 8 (6) : 209-213. |
| Thingstad T F, Lignell R. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol., 13 : 19-27. |
| Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acid. Res., 25 (24) : 4 876-4 882. |
| Venter J C, Remington K, Heidelberg J F, Halpern A L, Rusch D, Eisen J A, Wu D Y, Paulsen I, Nelson K E, Nelson W,Fouts D E, Levy S, Knap A H, Lomas M W, Nealson K,White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y H, Smith H O. 2004. |
| Environmental genome shotgun sequencing of the Sargasso Sea. Science, 304 (5667) : 66-74. |
| Waldor M K, Mekalanos J J. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science, 272 (5270) : 1 910-1 914. |
| Wang G H, Asakawa S, Kimura M. 2011. Spatial and temporal changes of cyanophage communities in paddy field soils as revealed by the capsid assembly protein gene g20.FEMS Microbiol. Ecol., 76 (2) : 352-359. |
| Wang K, Chen F. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol., 34 (2) : 105-116. |
| Wang M, Liang Y T, Bai X G, Jiang X J, Wang F, Qiao Q. 2010. Distribution of microbial populations and their relationship with environmental parameters in the coastal waters of Qingdao, China. Environ. Microbiol., 12 (7) : 1 926-1 939. |
| Waterbury J B, Watson S W, Valois F W, Franks D G. 1986.Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can. Bull.Fish. Aquat. Sci., 214: 71-120. |
| Weinbauer M G, Rassoulzadegan F. 2004. Are viruses driving microbial diversification and diversity? Environ.Microbiol., 6 (1) : 1-11. |
| Wichels A, Biel S S, Gelderblom H R, Brinkhoff T, Muyzer G,Schütt C. 1998. Bacteriophage diversity in the North Sea.Appl. Environ. Microbiol., 64 (11) : 4 128-4 133. |
| Wilhelm S W, Carberry M J, Eldridge M L, Poorvin L, Saxton M A, Doblin M A. 2006. Marine and freshwater cyanophages in a Laurentian Great Lake: evidence from infectivity assays and molecular analyses of g20 genes.Appl. Environ. Microbiol., 72 (7) : 4 957-4 963. |
| Wilhelm S W, Jeffrey W H, Dean A L, Meador J, Pakulski J D,Mitchell D L. 2003. UV radiation induced DNA damage in marine viruses along a latitudinal gradient in the southeastern Pacific Ocean. Aquat. Microb. Ecol., 31 (4) : 1-8. |
| Wilhelm S W, Suttle C A. 1999. Viruses and nutrient cycles in the sea-viruses play critical roles in the structure and function of aquatic food webs. Bioscience, 49 (10) : 781-788. |
| Wilhelm S W, Weinbauer M G, Suttle C A, Jeffrey W H. 1998.The role of sunlight in the removal and repair of viruses in the sea. Limnol. Oceanogr., 43 (1) : 586-592. |
| Wommack K E, Colwell R R. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev., 64 (1) : 69-114. |
| Yan Q, Wang M, Bai X, Sun J, Liang Y, Wang F. 2010. New phylogenetically distinct cyanophages found in the coastal Yellow Sea by Qingdao. Acta. Virologica., 54 (4) : 255-260. |
| Zhong Y, Chen F, Wilhelm S W, Poorvin L, Hodson R E. 2002.Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ.Microbiol., 68 (4) : 1 576-1 584. |
| Zwirglmaier K, Heywood J L, Chamberlain K, Woodward E M, Zubkov M V, Scanlan D J. 2007. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ. Microbiol., 9 (5) : 1 278-1 290." |
 2015, Vol. 33
2015, Vol. 33