Shanghai University
Article Information
- DU Guoying1, WU Feifei1, GUO Hao2, XUE Hongfan1, MAO Yunxiang1_L
- DNA barcode assessment of Ceramiales (Rhodophyta) in the intertidal zone of the northwestern Yellow Sea
- Chinese Journal of Oceanology and Limnology, 2015, 33(3): 685-695
- http://dx.doi.org/10.1007/s00343-015-4088-8
Article History
- Received Apr. 21, 2014;
- accepted in principle Jul. 8, 2014;
- accepted for publication Oct. 7, 2014
2 National Marine Environmental Monitoring Center, Dalian 100623, China
Ceramiales is one of the largest orders inRhodophyta(red algae)with currently up to 10families,408 genera,and 2 480 species registered inAlgaeBase(Guiry,2001; Guiry and Guiry,2014).Similar to most marine algae,with simplemorphologies,frequent convergence and highphenotypic variation in response to varyingenvironmental conditions,Ceramiales are alsodiffi cult to identify with certainty(De Jone et al.,1998; Lee et al.,2001; Lin et al.,2001; Saunders,2005,2008; Clarkston and Saunders,2010).
The DNA barcoding as a quick and accuratetechnique for species identifi cation was fi rst put forward in 1993(Arnot et al.,1993). This method hasbeen applied successfully in taxonomy and evolutionresearch on animals,microbes,and terrestrial plants(Hebert et al.,2003a,b; Yoo et al.,2006; Yancy et al.,2008; Hollingsworth et al.,2009). However,formarine algae,using a single gene does not meet therequirements of a barcode in terms of discriminatoryability and universality of PCR primers(Moritz and Cicero,2004). Until now,several markers for red macroalgae have been explored,including partialmitochondrial cytochrome c oxidase subunit I gene(COI),partial large-subunit rRNA gene(LSU),universal plastid amplicon(UPA),which is thedomain V of the plastid large subunit 23S ribosomalgene,and partial large(rbcL) and small(rbcS)subunits of ribulose-1,5-bisphosphate carboxylase(RuBisCO)gene,representing the cell nucleus,plastid,and mitochondrion genomes(Lee et al.,2001;Lin et al.,2001; Saunders,2005; Robba et al.,2006;Sherwood,2008; Sherwood et al.,2010b).
Each of these DNA markers has advantages and disadvantages in barcoding for red macroalgae. Forinstance,the UPA marker is easy to be amplifi ed and sequenced for most red algae(Sherwood and Presting,2007; Sherwood et al.,2010b),with appropriateinterspecifi c variation(1.1%–6.7%)on distinguishingmost species of red algae(Sherwood et al.,2010b;Zhao et al.,2012). The COI could be considered themost suitable DNA barcode for red algae because ofits remarkable ability to discriminate between closelyrelated species with large barcoding gaps(Saunders,2005,2008; Robba et al.,2006; Sherwood et al.,2010a,b). However,among the markers listed above,the COI is the most diffi cult to amplify and sequenceacross all red algae,even after seven different primercombinations were used(Clarkston and Saunders,2010; Sherwood et al.,2010a). It was deduced thatsequencing failures might be caused by theheterogeneity found with in species at positions nearthe 3' ends of the primers(Saunders,2008). Generally,rather than relying on a single marker,the combineduse of two or more markers has been recommended(Lin et al.,2001; Hall et al.,2010; Saunders and Kucera,2010).
In China,about 35 genera and 74 species ofCeramiales have been recorded,of which nearly 23 genera and 42 species have been reported to bedistributed along the northwestern coast of the YellowSea(Tseng,1984; Tseng et al.,2009). These recordsare sourced mostly from a taxonomic survey that wascarried out in the 1980s,with identifi cation based onmorphological and structural characteristics,as wellas reproductive features of Ceramiales. However,over the years since then,the biodiversity ofmacroalgae in coastal areas of China has decreaseddramatically as a result of anthropogenic impacts,such as aquaculture and recreation(Liu et al.,1999).Until now,only a few published studies of coastalRhodophyta from China have employed molecularapproaches(Zhao et al.,2012). Basic studies and information about macroalgal barcodes in coastalmacroalgae of China are needed to be established.This study aimed to get insights into the Ceramialesspecies of Rhodophyta,to generate baseline data ofmacroalgae,explore the potential cryptic or invasivespecies,and evaluate the biodiversity of coastalecosystem in the northwestern Yellow Sea. 2 MATERIAL AND METHOD 2.1 Sampling
From October 2011 to November 2012,142specimens of the order Ceramiales were collectedeach month from the intertidal zone of the northwesternYellow Sea during low tide(Fig. 1,Table 1).Morphological microstructure characteristics wereidentifi ed using an Olympus CX31 microscope(Olympus Co.,Japan)following Tseng et al.(2009),Zheng(2001),and Xia(2011). The fresh collectionswere cleared of epiphytes,and samples for molecularanalysis were frozen at -20°C. Representativespecimens have been preserved in the Algal Herbariumof Ocean University of China.
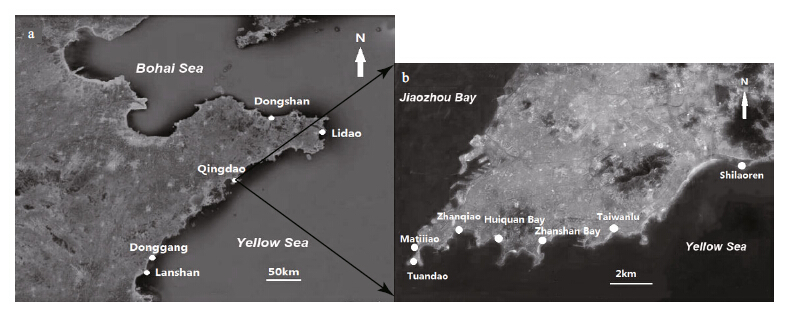 |
| Fig. 1 Sampling sites
a. northwestern coast of the Yellow Sea; b. coast of Qingdao. |
 |
DNA was extracted using a DNeasy Plant Kit(TIANGEN Biotech,Beijing,China)according to themanufacturer’s recommendation. LSU,UPA,and COIwere amplifi ed using the primers and ampli ficationprotocols described by Sherwood and Presting(2007),Sherwood et al.(2010b),and Saunders(2005). TheLSU sequence was amplifi ed using the followingprimers from Sherwood et al.(2010b): forwardnu28SF,5'-GGAATCCGCYAAG-GAGTGTG-3'(Tm=56.9°C) and reverse nu28SR,5'-TGCCGACTTCCCTTACCTGC-3'(Tm=59.7°C). The PCR cyclewas: 94°C for 2 min,40 cycles at 94°C for 20 s,55°Cfor 30 s,72°C for 50 s,and a fi nal extension at 72°Cfor 5 min(Sherwood et al.,2010b). The UPA sequencewas amplifi ed using the following primer pair:p23SrV_f1,5'-GGACAGAAAGACCCT-ATGAA-3' and p23SrV_r1,5'-TCAGCCTGTTATCCCTAGAG-3'(Sherwood and Presting,2007)The PCR cycle was:initial denaturation at 94°C for 2 min,followed by 35cycles at 94°C for 20 s,55°C for 30 s,and 72°C for 30s,and a fi nal extension at 72°C for 10 min. The COIsequence was amplifi ed using the following primers:GazF1,5'-TCAACAAATCATA-AAGATATTGG-3'(Saunders,2005) and R686,5'-CCACCWGMAGGATCAA-3'(Sherwood et al.,2010b). The PCR cyclewas: 94°C for 1.5 min,followed by 40 cycles at 94°Cfor 30 s,47°C for 40 s,72°C for 40 s,and a fi nalextension at 72°C for 5 min. The PCR ampli ficationswere performed with a Mycylcer thermal cycler(Bio-Rad,USA). Each 20 μL PCR solution consisted of10 μL PCR Mix,0.2 μL Taq(5 U/μL),1 μL of eachprimer(10 mmol/L),and 5.8 μL ddH2O. Thesequencing was performed by the BGI Biotech Co.Ltd.(Shenzhen,China).
Forward and reverse sequences of each samplewere aligned to obtain a complete and accuratesequence for each DNA barcode. The LSU,UPA,and COI sequences were then aligned using Clustal X1.83(UCD,Dublin,Irel and )(Thompson et al.,1994).After alignment,one voucher was selected to representa group of specimens with identical LSU,UPA,orCOI sequences for further cluster analyses. Specimensthat had identical LSU or UPA sequences but differentCOI sequences were also selected for further analyses.In addition,LSU,UPA,and COI sequences ofCeramiales species were downloaded from GenBankbased on the results of BLAST(Basic Local AlignmentSearch Tool)performing on NCBI(U.S. NationalCenter for Biotechnology Information,http://blast.ncbi.nlm.nih.gov/Blast.cgi; David and Medha,2007)against the nucleotide sequence database. Neighborjoining(NJ)trees were constructed based on themaximum-likelihood composite model in MEGA v.4(Tamura et al.,2007)using the Kimura 2-parametermethod that computes evolutionary distances(Kimura,1980). One thous and bootstrap replicateswere used to estimate the reliability of the branches.3 RESULT 3.1 Analyses of DNA markers
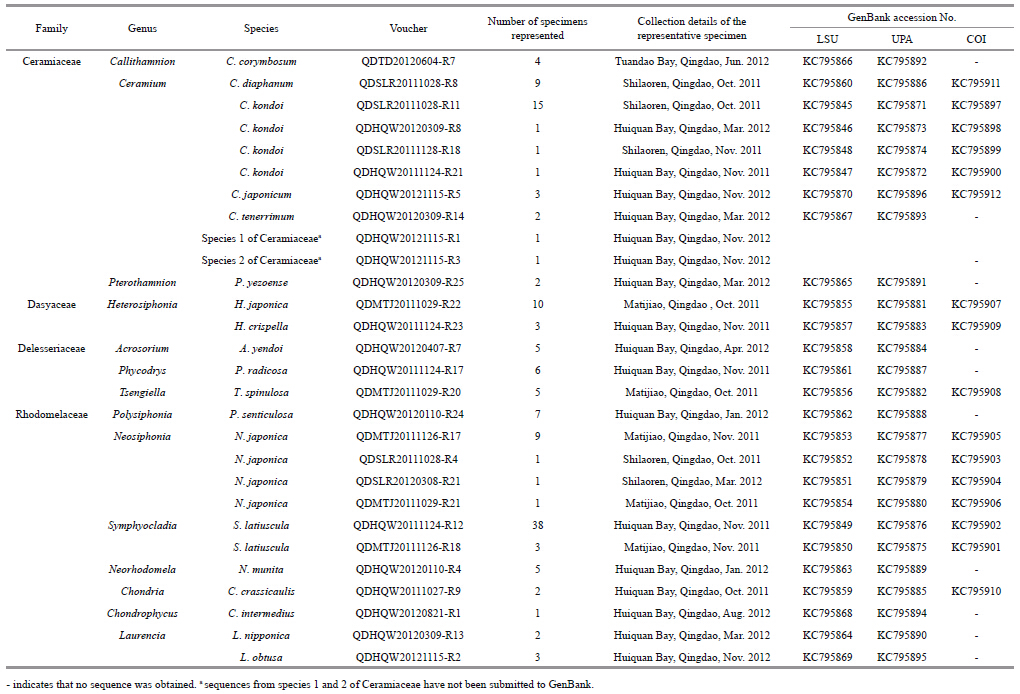
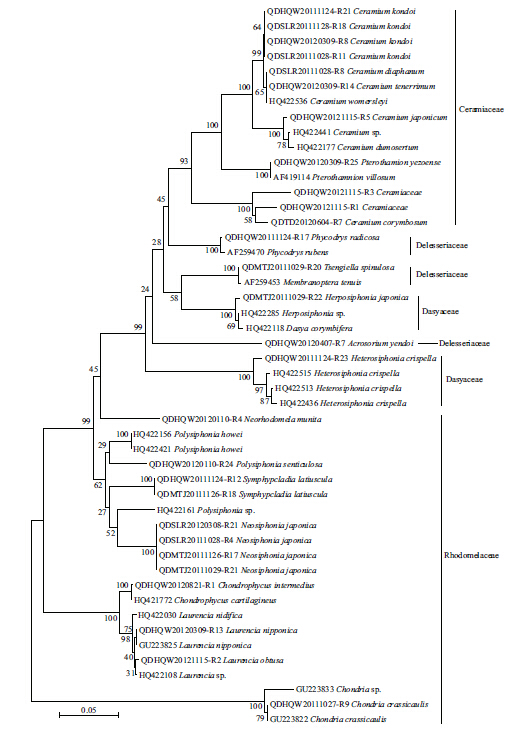
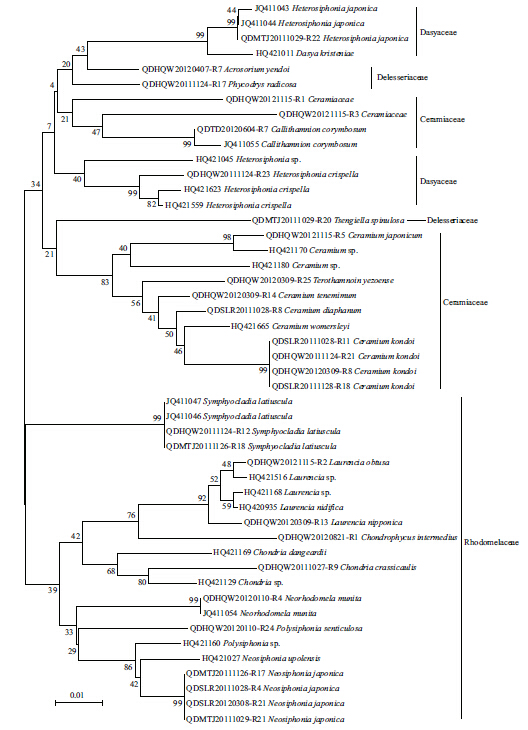
The LSU and UPA markers from all 142 specimenswere amplifi ed,and 137 sequences were obtained foreach of the markers. Only 83 of the COI markerscould be amplifi ed,out of which 39 sequences wereobtained. The ampli fication and sequencing successrate for LSU,UPA,and COI was 100% and 96.5%,100% and 96.5%,and 58.4% and 50.0%,respectively.The LSU and UPA sequences were obtained for allthe collected taxa,but no COI sequences wereobtained for 11 species(Table 1). Clustering treesbased on the specimen sequences and the sequencesfrom GenBank were constructed to illustrate thelevels of divergence with in and between species(Figs.2–4). The maximum intraspecific divergence(pairwise distances)was 0.025 for LSU,0.018 forUPA,and 0.152 3 for COI,all of which were insequences from Heterosiphonia crispella . Theminimum interspecifi c divergence(pairwisedistances)with in genus was 0 for LSU inPterothamnion and Ceramium,between P . yazoense and P . villosum,and among C . diaphanum,C .tenerrimum and C . womersleyi,0.003 for UPAbetween Laurencia sp. and Laurencia nidifica,and 0.029 6 for COI between Neosiphonia harveyi and Neosiphonia japonica . The maximum interspecifi cdivergence with in genus was 0.041 for LSU betweenPolysiphonia howei and Polysiphonia senticulosa,0.057 for UPA between Polysiphonia sp. and P .senticulosa,and 0.259 9 for COI between Ceramiumkondoi and Ceramium japonicum . When the overallpairwise distances were compared it was clear that theCOI had the highest values among the three markers.
 |
| Fig. 2 Neighbor-joining tree of LSU sequences from Ceramiales
The sequences from the specimens collected for this study are those serial numbers with “-Rxx”; the other sequences are from GenBank. Bootstrap valueswere obtained from 1 000 replications(values below 50% are not shown). |
 |
| Fig. 3 Neighbor-joining tree of UPA sequences from Ceramiales
The sequences from the specimens collected for this study are those serial numbers with “-Rxx”; the other sequences are from GenBank. Bootstrap valueswere obtained from 1 000 replications(values below 50% are not shown). |
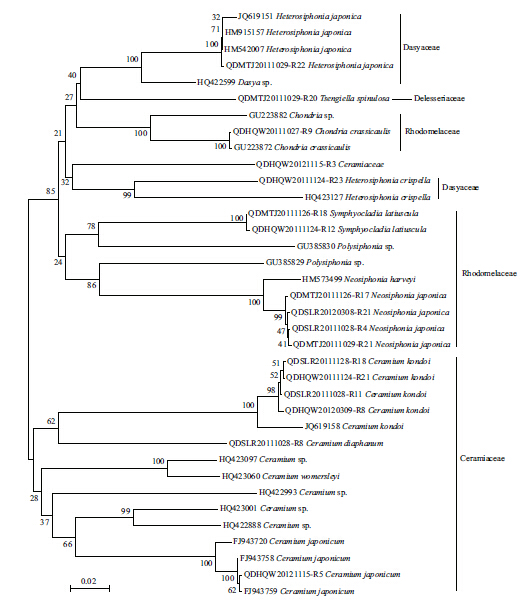 |
| Fig. 4 Neighbor-joining tree of COI sequences from Ceramiales
The sequences from the specimens collected for this study are those serial numbers with “-Rxx”; the other sequences are from GenBank. Bootstrap valueswere obtained from 1 000 replications(values below 50% are not shown). |
Our collection covers 21 species,14 genera,and four families of Ceramiales. Eleven of the speciesbelong to 11 genera(Table 1); four were assigned togenus Ceramium,two were Heterosiphonia,and twowere Laurencia . The remaining two species(species1 and 2)were assigned only to the family Ceramiaceae (Table 1). No discrepancies were detected in the threemarker sequences with in most of the species; theexceptions were for three species C . kondoi,Symphyocladia latiuscula,and N . japonica in which a1–3 bp divergence was found in the COI sequences.
In the NJ trees for LSU,UPA,and COI,a proportionof the branch supports(23%,41%,and 36%,respectively)were lower than 50%. Not all the genera from same family were clustered together,except forCeramiaceae and Rhodomelaceae in LSU Tree,Rhodomelaceae in UPA tree,and Ceramiaceae in COItree(Figs.2–4). In all the NJ trees of LSU,UPA,and COI,Heterosiphonia japonica fi rst converged withDasya spp. and were distant from the otherHeterosiphonia spp. At the genus and species levels,the LSU marker exhibited low resolution,especially between Polysiphonia and Neosiphonia,but alsoamong Ceramium diaphanum,Ceramium tenerrimum,and Ceramium womersleyi . On the contrary,the UPA and COI trees could discriminate among genera and species,and the COI tree also showed sequencedivergence with in species such as C . japonicum,C .kondoi,and N . japonica(Fig. 4).
Nine of the species in our collection had little or nosequence divergence with their correspondingsequences from GenBank; for example,the H .japonica were identical in both UPA and COI,theChondria crassicaulis were consistent in LSU and COI,S . latiuscula congruent in UPA and only had 1bpdifference on COI. The C . japonicum presented littledistance of 0.001 with GenBank sequence FJ943759on COI,the Neorhodomela munita UPA had nodistance with GenBank sequence JQ411054,and alsothe Laurencia nipponica LSU had no difference withGenBank sequence GU223825. The Callithamnioncorymbosum had a distance of 0.045 on UPA fromGenBank sequence JQ411055. The sequences of C .kondoi diverged from GenBank COI sequenceJQ619158 by 0.033–0.037,and H . crispella hadsequence distances to same species from GenBank by0.019,0.018,and 0.157 on LSU(HQ422515),UPA(HQ421559) and COI(HQ423127),respectively,with strong support by 99 and 100 bootstraps in theirclustering trees(Figs.2–4).
Ten species and species1 and species 2 ofCeramiaceae failed to fi nd matches by sequencealignment,due to lack of corresponding genesequences in GenBank or BOLD,respectively.4 DISCUSSION
4 DISCUSSIONThe LSU,UPA,and COI markers identifi ed in thepresent study showed consistent applicability and capability of species discrimination with the resultsfrom previous studies(Saunders,2005,2008; Robba et al.,2006; Sherwood and Presting,2007; Sherwood et al.,2010a,b). That is,LSU and UPA were amplifi ed and sequenced easier than COI across all theCeramiales species,but the LSU marker could not beused to discriminate at the species level. LSU,forexample,could not distinguish the Ceramium speciesC . womersleyi,C . diaphanum,and C . tenerrimumbecause there was no sequence divergence amongthem. On the other h and ,COI had better discriminatingability than LSU and UPA,with the least conservedindicated by large divergences or long branches. Inthis study,three species confi rmed their classifi cationby molecular accordance on COI. Besides,the COI sequence diversity was found at intraspecific level inC . kondoi,N . japonica and S . latiuscula,supportingthe ability of this gene in identifying genetic diversitywith in species(Kim et al.,2010).
Theoretically,DNA barcoding has obviousadvantages for species identifi cation and for geneticdiversity evaluation,not only because of itsindependence from morphological features,whichusually need accumulative experience indistinguishing species with intricate convergence and plastic phenotype,but also because it does not rely onreproductive structures or life cycles(Xiao et al.,2004; Saunders 2005,2008; Evans et al.,2007;Sherwood et al.,2010a,b). Even with in a species,high diversity and various phylogenetic relationshipshave been discovered through DNA barcoding(Sherwood et al.,2011; Kim et al.,2012). In thepresent study,Heterosiphonia species were separatedby other family and failed to cluster together in allthree NJ trees,suggesting that this genus has largedivergence in the nucleotide sequences. In all threeNJ trees,H . japonica converged fi rst with Dasya spp. and was distant from other Heterosiphonia spp.,corroborating previous deductions(de Jong et al.,1998; Choi,2001; Choi et al.,2002)that the Dasyaceaefamily is polyphyletic and that H . japonica hasaffi nities to the genus Dasya indicated by phylogeneticanalyses of anatomical and nuclear SSU rDNAsequence. In our study,only six species(three by COI and four by UPA)could be confi rmed by sequencematching. On the other h and ,because short DNAmarkers were used,the phylogenetic relationships and branch support are not suitable to be assessed(Sherwood et al.,2010b). In our study,because of lowsequences achievement for COI,the phylogeneticanalysis of concatenated three markers failed to becarried out. The analysis based on the concatenatedLSU and UPA(less than 1 000 bp)also showed lowsupport with bootstrap values that were mostly lessthan 70%(data no shown). Therefore,only thepatterns of diversity for species and groups of closelyrelated species could be evaluated.
Cryptic or invasive species have often beenrevealed by DNA barcoding(Robba et al.,2006;Saunders,2008,2009; Rueness,2010; Sherwood et al.,2010a,b). Three specimens(represented byQDHQW20111124-R23)that were hard to identifywithout reproductive structures,even theirmorphological structure are similar to H . crispella .By analyses of three NJ trees,we presumed that theymight belong to H . crispella based on the strong support of their sequence clustered together withGenBank sequences of H . crispella,although thesequence distance is large on COI as 0.157,but closeron UPA as 0.018. This species has not been recordedpreviously in China(Tseng,1983; Zheng,2001;Tseng et al.,2009; Xia,2011),which implies that itmight be a cryptic or invasive species in theinvestigated region.
On the other h and ,the shortage of related sequencesin the available databases seriously impeded theapplication of DNA barcoding for speciesidentifi cation. In this study,12 species failed to fi ndmatched sequences from GenBank or BOLD. At themeantime,it should be noted that using GenBanksequences to identify species should be treated withcaution for two reasons: one,accurate identifi cationof species cannot be guaranteed(Harris,2003;Vilgalys,2003); and two,the quality assurance of thesequences in GenBank is less rigorous. And somesequences may contain large gaps or many ambiguoussites that decrease the strength of multiple sequenceanalyses for the purposes of species identifi cation and may infl ate the perceived levels of species diversity(Le Gall and Saunders,2010).
Therefore,simply to depend on molecularidentifi cation is not feasible,and DNA barcodingshould base on solidly traditional morphologicalidentifi cation(Bensasson et al.,2001; Hebert et al.,2003a; Schindel and Miller,2005; Witt et al.,2006;Maggs et al.,2007). At present,a st and ard barcodemarker that is suitable for all orders or families hasnot been established,and a combination of severalDNA markers is still necessary for speciesidentifi cation(Saunders,2008; Sherwood et al.,2010a). For DNA barcoding of red algae,such asCeramiales,we suggest that UPA and COI could beapplied complementarily,as recommended bySherwood et al.(2010b).
A total of 14 genera and 21 species of Ceramialeswere collected in our 1-year monthly survey. In their,books,Tseng(1984) and Tseng et al.(2009)haverecorded nearly 23 genera and 42 species that aredistributed along the northwestern coast of the YellowSea. Based on investigations conducted in May 1998 and May 1999,Liu et al.(1999)reported 23 genera and 36 species of red algae(but only 12 species ofCeramiales)in the Qingdao intertidal zone. FromAugust 2004 to May 2005,Fu et al.(2009)carried outmonthly monitoring on benthic macroalgae in rockyintertidal zones of Qingdao and reported a total 13species of Ceramiales. The species diversity explored in the present study is lower than in the records ofTseng(1984) and Tseng et al.(2009),but higher thanthe records of Liu et al.(1999) and Fu et al.(2009).The investigated area and period in our study arerelatively less than those of Tseng(1984) and Tseng etal.(2009),but more expansive than those of Liu et al.(1999) and Fu et al.(2009). Thus,it is hard to deducethat the biodiversity in these regions has decreaseddramatically because of the anthropogenic impact asconcluded by Liu et al.(1999). Further studies need tobe carried out to fully investigate the biodiversity ofCeramiales and other macroalgae species in thisregion.5 CONCLUSION
In this study,142 specimens that covered 21species,14 genera,and four families were assessed byDNA barcoding. The results indicated that in theintertidal zone of the northwestern Yellow Sea,therewas high diversity between the Ceramiales species,while,with in a species,were relatively conservative.This study also corroborated previous reports thatDNA barcoding is a quick and helpful technique fortaxonomy and for uncovering invasive or crypticspecies. However,because short DNA markers wereused,it was hard to evaluate phylogenetic relationshipsat higher taxa levels. Considering the shortage orinaccurate information of DNA barcodes formacroalgae that is available,at present,DNAbarcoding for species identifi cation still needs to beused in combination with traditional morphologicalmethods.
| Arnot D E, Roper C, Bayoumi R A L.1993. Digital codes from hypervariable tandemly repeated DNA sequences in the Plasmodium falciparum circumsporozoite gene can genetically barcode isolates. Mol. B iochem. P arasit., 61 (1): 15-24. |
| Bensasson D, Zhang D X, Hartl D L, Hewitt G M. 2001.Mitochondrial pseudogenes: evolution's misplaced witnesses. Trends. Ecol. Evol., 16 : 314-321. |
| Chase M W, Salamin N, Wilkinson M, Dunwell J M,Kesanakurthi R P, Haidar N, Savolainen V. 2005. Land plants and DNA barcodes: short-term and long-term goals. Philos. T rans. R. Soc. Lond. B Biol. Sci., 360 (1462): 1 889-1 895. |
| Choi H G, Kraft G T, Lee I K, Sauders G W. 2002. Phylogenetic analyses of anatomical and nuclear SSU rDNA sequence data indicate that the Dasyaceae and Delesseriaceae (Ceramiales, Rhodophyta) are polyphyletic. Eur. J.Phycol., 37 : 551-569. |
| Choi H G. 2001. Morphology and reproduction of Heterosiphonia pulchra and H. japonica (Ceramiales,Rhodophyta). Algae, 16 : 387-409. |
| Clarkston B E, Saunders G W. 2010. A comparison of two DNA barcode markers for species discrimination in the red algal family Kallymeniaceae (Gigartinales,Florideophyceae), with a description of Euthora timburtonii sp. nov. Botany, 88 : 119-131. |
| Conklin K Y, Kurihara A, Sherwood A R. 2009. A molecular method for identification of the morphologically plastic invasive algal species Eucheuma denticulatum and Kappaphycus spp. (Rhodophyta, Gigartinales) in Hawaii.J. Appl. Phycol., 21 : 691-699. |
| David W, Medha B. 2007. Chapter 9: BLAST QuickStart. In :Nicholas B H ed. Comparative Genomics Volumes 1 and 2. Methods in Molecular Biology. Humana Press, Totowa,New Jersey, USA. p.395-396. |
| De Jong Y S D M, Vanderwurff A W G, Stam W T, Olsen J L. 1998. Studies on Dasyaceae 3. Towards a phylogeny of the Dasyaceae (Ceramiales, Rhodophyta) based on comparative rbcL gene sequences and morphology. Eur.J. P hycol., 33 : 187-201. |
| Evans K M, Wortley A H, Mann D G. 2007. An assessment of potential diatom “Barcode” Genes (cox 1, rbc L, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist., 158 : 349-364. |
| Fu J P, Zeng X Q, Wang C Y. 2009. Study on community of benthic macroalgae in the rocky intertidal zone of Qingdao. J. Ocean Univ. China, 39 (Sup.): 25-31. (in Chinese with English abstract) |
| Guiry M D, Guiry G M. 2014. AlgaeBase. World-wide electronic publication, National University of Ireland,Galway. http://www.algaebase.org. Accessed on 2014-07-10. |
| Guiry M D. 2001. Macroalgae of Rhodophycota, Phaeophycota,Chlorophycota, and two genera of Xanthophycota, I n :Costello M J ed. European Register of Marine Species: A Check-list of the Marine Species in Europe and A Bibliography of Guides to Their Identification. Collection Patrimoines Naturels, 50 : 20-38. |
| Hajibabaei M, Janzen D H, Burns J M, Hallwachs W, Hebert P D N. 2006. DNA barcodes distinguish species of tropical Lepidoptera. P. Nat l. A Sci. India A, 103 : 968-971. |
| Hall J D, FučíkovÁ K L O C, Lewis L A, Karol K G. 2010. An assessment of proposed DNA barcodes in freshwater green algae. Cryptogamie Algol., 31 (4): 529-555. |
| Harris J D. 2003. Can you bank on GenBank? Trends. Ecol.Evol., 18 : 317-319. |
| Hebert P D N, Cywinska A, Ball S L, deWaard J R. 2003a.Biological identifications through DNA barcodes. P. Roy.Soc. B-B iol. S ci., 270 : 313-322. |
| Hebert P D N, Ratnasingham S, deWaard J R. 2003b. Barcoding animal life: cytochrome c oxidase subunit I divergences among closely related species. P. Roy. Soc. B-Biol. Sci., 270 : S96-S99. |
| Hebert P D N, Stoeckle M Y, Zemlak T S, Francis C M. 2004.Identification of birds through DNA barcodes. PloS Biol., 2 : 1 657-1 663. |
| Hollingsworth P M, Forrest L L, Spouge J L. 2009. A DNA barcode for land plants. P. Natl. Acad. Sci. USA., 106 (31): 12 794-12 797. |
| Kim M S, Yang M Y, Cho G Y. 2010. Applying DNA barcoding to Korean Gracilariaceae (Rhodophyta). CryptogamieAlgol., 31 (4): 387-401. |
| Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol., 16 : 111-120. |
| Le Gall L, Saunders G W. 2010. DNA barcoding is a powerful tool to uncover algal diversity: a case study of the Phyllophoraceae (Gigartinales, Rhodophyta) in the Canadian flora. J. Phycol., 46 : 374-389. |
| Lee S R, Oak J H, Suh Y, Lee I K. 2001. Phylogenetic utility of rbc S sequences: an example from Antithamnion and related genera (Ceramiaceae, Rhodophyta). J. Phycol., 37 (6): 1 083-1 090. |
| Lin S M, Fredericq S, Hommersand M H. 2001. Systematics of the Delesseriaceae (Ceramiales, Rhodophyta) based on large subunit rDNA and rbcL sequences, including the Phycodryoideae, subfam. nov. J. Phycol., 37 (5): 881-899. |
| Liu D Y, Wang Z Y, Sun J, Huang Z Y, Qian S B. 1999. Study of the benthic algae in the littoral of Qingdao coast. Trans.Oceanol. Limnol., 3 : 35-40. (in Chinese) |
| Maggs C A, Verbruggen H, De Clerck O. 2007. 6 Molecular systematics of red algae: building future structures on firm foundations. I n : Brodie J ed. Unravelling the Algae: the Past, Present, and Future of Algal Systematics, The Systematics Association Special Volume Series, 75. CRC Press: Boca Raton. p.103-121. |
| Moritz C, Cicero C. 2004. DNA barcoding: promise and pitfalls. PloS Biol., 2 : 1 529-1 531. |
| Presting G G. 2006. Identification of conserved regions in the plastid genome implications for DNA barcoding and biological function. Can. J. Bot., 84 : 1 434-1 443. |
| Robba L, Russell S, Baker G, Brodie J. 2006. Assessing the use of the mitochondrial COX I marker for use in DNA barcoding of red algae (Rhodophyta). Am. J. Bot., 93 (8): 1 101-1 108. |
| Rueness J. 2010. DNA barcoding of select freshwater and marine red algae (Rhodophyta). Cryptogamie Algol., 31 (4): 377-386. |
| Saunders G W, Hommersand M H. 2004. Assessing red algal supraordinal diversity and taxonomy in the context of contemporary systematic data. Am. J. Bot., 91 : 1 494-1 507. |
| Saunders G W, Kucera H. 2010. An evaluation of rbcL, tufA,UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie Algol., 31 (4): 487-528. |
| Saunders G W. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future application. Philos. T rans. R. Soc. Lond. B Biol.Sci., 360 : 1 879-1 888. |
| Saunders G W. 2008. A DNA barcode examination of the red algal family Dumontiaceae in Canadian waters reveals substantial cryptic species diversity. 1. The foliose Dilsea-Neodilsea complex and Weeksia. Bot., 86 : 773-789. |
| Saunders G W. 2009. Routine DNA barcoding of Canadian Gracilariales (Rhodophyta) reveals the invasive species Gracilaria vermiculophylla in British Columbia. Mol.Ecol. Res., 9 : 140-150. |
| Schindel D E, Miller S E. 2005. DNA barcoding a useful tool for taxonomists. Nature, 435 : 17. |
| Sherwood A R, Chan Y, Presting G G. 2008. Application of universal plastid primers to environmental sampling of a Hawaiian stream periphyton community. Mol. Ecol.Res our., 8 : 1 011-1 014. |
| Sherwood A R, Kurihara A, Conklin K Y, Sauvage T, Presting G G. 2010b. The Hawaiian Rhodophyta Biodiversity urvey (2006-2010): a summary of principal findings.BMC Plant Biol., 258 (10): 1 471-2 229. |
| Sherwood A R, Kurihara A, Conklin K Y. 2011. Molecular diversity of Amansieae (Ceramiales, Rhodophyta) from the Hawaiian Islands: a multi-marker assessment reveals high diversity with in Amansia glomerata. Phycol. Res., 59 : 16-23. |
| Sherwood A R, Presting G G. 2007. Universal primers amplify a 23 S rDNA plastid marker in eukaryotic algae and cyanobacteria. J. Phycol., 43 : 605-608. |
| Sherwood A R, Sauvage T, Kurihara A, Conklin K Y, Presting G G. 2010a. A comparative analysis of COI, LSU and UPA marker data for the Hawaiian florideophyte Rhodophyta: implications for DNA barcoding of red algae. C ryptogamie Algol., 15 : 451-465. |
| Tamura K, Dudley J, Nei M, Kumar M S. 2007. MEGA4:Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol., 24 : 1 596-1 599. |
| Thompson J D, Higgins D G, Gibson T J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res., 22 : 4 673-4 680. |
| Tseng C K, Xia B M, Zhou X T. 2009. Seaweeds in Yellow Sea and the Bohai Sea of China. Science Press, Beijing, China. 254p. (in Chinese) |
| Tseng C K.1984. Common Seaweeds of China. Science Press,Beijing, China. 314p. |
| Vijayan K, Taou C. 2010. DNA barcoding in plants: taxonomy in a new perspective. Curr. Sci. India, 99 (11): 1 530-1 541. |
| Vilgalys R. 2003. Taxonomic misidentification in public DNA databases. New Phytol., 160 : 4-5. |
| Witt J D S, Therloff D L, Hebert P D N. 2006. DNA barcoding reveals extraordinary cryptic diversity in an amphipod genus: implications for desert spring conservation. Mol.Ecol., 15 : 3 073-3 082. |
| Xia B M. 2011. Chinese Seaweed: Rhodophyta. 2(7). Science Press, Beijing, China. p.212. (in Chinese) |
| Xiao J H, Xiao H, Huang D W. 2004. DNA barcoding; new approach of biological taxonomy. Curr. Zool., 50 (5): 852-855. |
| Yancy H F, Zemlak T S, Mason J A, Washington J D, Tenge B J, Nguyen N L T, Barnett J D, Savary W E, Hill W E Moore M M, Fry F S, Randolph S C, Rogers P L, Hebert P D N. 2008. Potential use of DNA barcodes in regulatory science: applications of the regulatory fish encyclopedia.J. Food Protect., 71 : 210-217. |
| Yoo H S, Eah J Y, Kim J S, Kim Y J, Min M S, Paek W K, Lee H, Kim C B. 2006. DNA barcoding Korean birds. Mol. Cells, 22 : 323-327. |
| Zhao X B, Peng S J, Shan T F, Liu F. 2012. Applications of three DNA barcodes in assorting intertidal red macroalgal flora in Qingdao, China. J. Ocean Univ. China, 12 : 139-145. |
| Zheng B L. 2001. Chinese Seaweed: Rhodophyta. Vol. 2(6).Science Press, Beijing, China. p.159. (in Chinese) |
 2015, Vol. 33
2015, Vol. 33


