Shanghai University
Article Information
- ZHANG Hao(张好), LI Fuchao(李富超), CHEN Huaxin(陈华新), ZHAO Jin(赵瑾), YAN Jinfei(闫晋飞), JIANG Peng(姜鹏), LI Ronggui(李荣贵), ZHU Baoli(朱宝利)
- Cloning, expression and characterization of a novel esterase from a South China Sea sediment metagenome
- Chinese Journal of Oceanology and Limnology, 2015, 33(4): 819-827
- http://dx.doi.org/10.1007/s00343-015-4170-2
Article History
- Received Jun. 12, 2014
- accepted in principle Aug. 2, 2014;
- accepted for publication Aug. 13, 2014
2 Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China
3 Third Institute of Oceanography, State Oceanic Administration, Xiamen 361005, China
4 Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China
Lipolytic enzymes, including esterase(EC 3.1.1.1) and lipase(EC 3.1.1.3), are a group of hydrolases that catalyze the cleavage and formation of ester bonds. Most lipolytic enzymes have a wide substrate tolerance, high stability in organic solvents, and no specific cofactor requirement, which are characteristics that make lipolytic enzymes easy to catalyze. Lipolytic enzymes consequently have potential applications in laundry, food engineering, pharmaceutical and biodiesel production(Bornscheuer, 2002).
Lipolytic enzymes are typically derived from animals, plants and microorganisms. Nevertheless, fewer than 1% of marine microorganisms can be cultivated in the laboratory by conventional methods(Amann et al., 1995). Metagenomics is widely used for functional gene screening of uncultured microorganisms(Handelsman et al., 1998; Schmeisser et al., 2007). Lipolytic enzymes have been identified using metagenomic libraries from pond water(Ranjan et al., 2005), soils(Lämmle et al., 2007), activated sludge(Roh and Villatte, 2008), plant rhizosphere soil(Lee et al., 2010)sheep rumen(Bayer et al., 2010) and various marine environments such as tidal flat sediments(Lee et al., 2006), seawater(Chu et al., 2008), intertidal flat sediments(Kim et al., 2009) and deep-sea sediments(Park et al., 2007; Jeon et al., 2009; Hu et al., 2010; Fu et al., 2011; Jiang et al., 2012).
Nearly 71% of the earth’s surface is covered by ocean. The marine environment is extremely diverse, with features such as cold seeps, hydrothermal vents, abyssal plains, oceanic trenches, seamounts, oceanic volcanoes and deep-sea whale falls, as well as varied conditions of pressure, salinity, temperature, nutrient composition and light. Marine microorganisms are a huge untapped source of lipolytic enzymes, especially esterase, with extreme marine environments thus constituting a vast pool of novel lipolytic biocatalysts(Kennedy et al., 2008). As a consequence of this diverse marine environment, various lipolytic enzymes have distinct characteristics, such as thermostability, low temperature activity and heavy metal tolerance.
In our study, we designed primers to directly screen the esterase genes from the metagenomic DNA of South China Sea deep-sea sediments. The esterase gene scsEst01 was cloned and co-expressed in Escherichia coli BL21(DE3)with the chaperone dnaK-dnaJ-grpE. Our results reveal that scsEst01 is a novel meso-thermophilic alkaline esterase.
2 MATERIAL AND METHOD 2.1 Metagenomic DNA extraction and esterase gene cloningDeep-sea sediments were collected from the northern portion(114°46.684 3'E, 19°11.674 0'N)of the South China Sea at a water depth of 1 394 m during the cruise aboard the R/V Ke Xue Yi Hao. This sampling area is generally considered to be a methaneenriched area. Metagenomic DNA was extracted from 0.5 g sediment samples using a PowerSoil DNA Isolation kit(MoBio, West Carlsbad, CA, USA) following the manufacturer’s instructions.
An esterase gene was amplified using metagenomic DNA as a template with the following primers: 5'-GGAATTCCATATGGCCAGCCAGCCAGCACCTTC- 3'(Nde I site underlined) and 5'-CCCAAGCTTTTAGGCTGCCTGCCGCCTCCGGGA-3'(Hind III site underlined). The amplified esterase gene was ligated into a pMD18-T vector(TaKaRa, Dalian, China) and transformed into E.coli Top10. The pMD18-TscsEst01 positive clone was sequenced at Shanghai Majorbio Bio-pharm Technology Co.(Shanghai, China).
2.2 ScsEst01 sequence analysisThe scsEst01 gene was annotated by a BLASTX search of the NCBI non-redundant protein sequence (nr)database. Multiple alignments between amino acid sequences of lipolytic enzymes were performed using CLUSTAL X(Thompson et al., 1997), with conserved blocks across the aligned sequences visualized with DNAMAN software. A phylogenetic tree was constructed by the neighbor-joining method using MEGA v4.0 software(Tamura et al., 2007).
2.3 Overexpression with chaperones and purification of scsEst01The scsEst01 gene was digested by Nde I and Hind III(MBI Fermentas, Ottawa, Canada)from pMD18- T-scsEst01 and cloned into a pET-28a expression vector(Novagen, Billerica, MA, USA)digested with the same enzymes. The pET-28a-scsEst01 recombinant plasmid was transformed into E.coli Transetta(DE3)chemically competent cells. After induction with 0.5 mmol/L isopropyl-β-Dthiogalactoside(IPTG)at different temperatures(16, 25, and 28°C)for 8–14 h, the target protein was expressed. Under each of these induction conditions, however, the recombinant protein was generated as an inactive inclusion body.
Chaperone plasmid sets(Takara)pTf16, pKJE7, pG-Tf2, pGro7 and pG-KJE8 combined individually with pET-28a-scsEst01 were co-transformed into E. coli BL21(DE3). The transformants were inoculated in Luria-Bertani(LB)medium with 20μg/mL chloramphenicol, 50 μg/mL kanamycin, 0.5 mg/mL L-arabinose and /or 5 ng/mL tetracycline at 37°C, and then induced for 8 h with 0.5 mmol/L IPTG at 25°C. The crude lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis(SDSPAGE).
The chaperone plasmid set pKJE7(coding dnaKdnaJ-grpE chaperones)with pET-28a-scsEst01 was selected for medium-scale fermentation. Recombinant E.coli cells were cultured in 2.5 L of LB medium containing 20 μg/mL chloramphenicol, 50 μg/mL kanamycin and 0.5 mg/mL L-arabinose at 37°C with 200 r/min shaking. When the OD600 reached 0.5, the cultures were induced with 0.5 mmol/L IPTG overnight at 25°C. Cells were harvested by centrifugation at 8 000×g for 20 mmol/L sodium chloride and 20 mmol/L imidazole, pH 7.4) and disrupted by sonication on ice. The cell debris was removed by centrifugation at 12 000×g for 20 min at 4°C. The supernatant was loaded onto an immobilized metal ion affinity column containing chelating Sepharose(GE Healthcare Bio-Sciences, Uppsala, Sweden)charged with Ni2+, which was preequilibrated with breaking buffer. Host proteins and chaperones were removed with washing buffer (20 mmol/L sodium phosphate, 500 mmol/L sodium chloride and 50 mmol/L imidazole, pH 7.4). The target protein was eluted with elution buffer (20 mmol/L sodium phosphate, 500 mmol/L sodium chloride and 500 mmol/L imidazole, pH 7.4). The peak fraction was pooled and imidazole was removed using a Sephadex G25 column(GE Healthcare BioSciences), and the proteins were then subjected to 12% SDS-PAGE analysis.
2.4 Characterization of the esterase scsEst01ScsEst01 protein concentration was measured by the method of Bradford(1976). Enzyme activity was determined using p-nitrophenol(p-NP)butyrate as a substrate. The st and ard reaction mixture contained 0.04 mL of 10 mmol/L p-NP butyrate in acetonitrile, 3.92 mL of 50 mmol/L Tris-HCl buffer(pH 7.5) and 0.04 mL enzyme in a final volume of 4 mL(Park et al., 2007). Enzyme activity was determined at 30°C by measuring the absorbance of p-NP released at 405 nm. One unit of esterase activity was defined as the amount of enzyme needed to release 1 μmol p-NP per min(Park et al., 2007).
The optimum temperature for protein activity was determined at 25–50°C. The effect of pH on enzyme activity of scsEst01 was measured over a pH range of 5.0–9.2 with different pH buffers, namely 100 mmol/L citrate buffer(pH 5.0–6.5), 100 mmol/L potassium phosphate buffer(pH 6.5–7.5), 100 mmol/L Tris-HCl buffer(pH 7.5–9.0), and 50 mmol/L CHES buffer (pH 9.0–9.2)(Jiang et al., 2012). Thermostability was examined by measuring residual enzyme activity after incubation of scsEst01 for 30 min at -20, 0, 10, 20, 30, 40, 50, and 60°C. After mixing esterase scsEst01 with equal volumes of various pH phosphate buffers(pH range of 5.0–9.2)at 4°C overnight, activity of the enzyme using p-NP butyrate as the substrate was tested at 35°C in 100 mmol/L Tris-HCl buffer(pH 8.0).
The effects of metal ions(Ni+, Co2+, Fe2+, Ca2+, Cu2+, Zn2+, Mg2+ and Mn2+)were determined at a final concentration of 10 mmol/L. The effects of detergents 1% Tween 20, Tween 80, TritonX-100, SDS, and CTAB were also examined. The effect of organic solvents was determined using methanol, ethanol, isopropanol, acetone, dimethylsulfoxide(DMSO), and dimethylformamide(DMF)at afinal concentration of 15%. After pre-incubation for 30 min with each reagent, all tests were performed using st and ard assays, with enzyme activity without additives defined as 100% activity.
3 RESULT 3.1 ScsEst01 gene cloning and sequence analysisThe scsEst01 gene was cloned from sediment metagenomic DNA and sequenced as 921 bp encoding 307 amino acids. A BLASTX comparison of the translated protein sequence showed less than 90% identity at the amino acid level with proteins from other uncultured bacteria based on metagenomic screening. A phylogenetic tree was constructed using nine matched sequences. As shown in Fig. 1, scsEst01 was found to be in a sister group relationship with FLS15(ACL67849; 89% identity)within a group of esterases from South China Sea sediment(Hu et al., 2010).
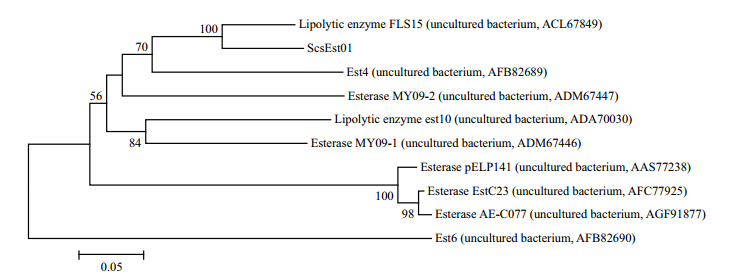 |
| Fig. 1 Phylogenetic tree of esterase amino acid sequences generated with MEGA v4.0 using the neighbor-joining method Bootstrap support values ≥50% based on 1 000 replicates are shown at nodes. |
A multiple sequence alignment of amino acid sequences of these esterases is shown in Fig. 2. All aligned esterases contained the lipase-conserved catalytic triad residues Asp, His and the catalytic nucleophile Ser in the consensus pentapeptide GDSAG(G-X-S-X-G type). These results place the esterases into family IV, a family whose members display a striking amino-acid sequence similarity to mammalian hormone-sensitive lipase(HSL). The HGGG motif may serve as the oxyanion hole of family IV lipases.
 |
| Fig. 2 Multiple alignment of esterase amino acid sequences including conserved blocks The consensus pentapeptide G-X-S-X-G block(boxed)was predicted to the catalytic triad. The HGGG motif may be the family-IV lipase oxyanion hole. |
The nucleotide sequence of the scsEst01 gene has been deposited in GenBank(accession number KJ935031).
3.2 Expression and purification of scsEst01Although recombinant esterase scsEst01 was overproduced in E.coli Transetta(DE3), most of the target protein was expressed as inclusion bodies under IPTG induction at different temperatures(16, 25 and 28°C) for 8–14 h. To improve the solubility of scsEst01, we unsuccessfully tested various expression-related factors, such as culture temperature, induction temperature and IPTG concentration. Chaperone plasmid sets pTf16, pKJE7, pG-Tf2, pGro7 and pGKJE8 were then used for co-expression with pET-28a-scsEst01 in E.coli BL21(DE3). As shown in Fig. 3, the chaperone plasmid pKJE7(coding for dnaK-dnaJ-grpE chaperones)performed well to form soluble recombinant esterase scsEst01.
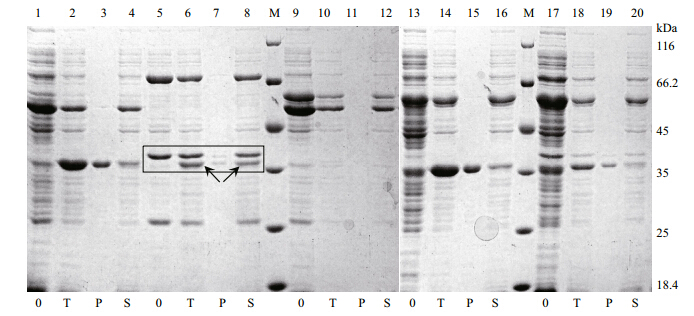 |
| Fig. 3 Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of crude lysates of E.coli BL21(DE3)carrying scsEst01
co-expressed with chaperones 0: crude lysate without induction; T: crude lysate after induction; P: precipitate of induced crude lysate after centrifugation; S: supernatant of induced crude lysate after centrifugation. Lanes 1–4: pTf16 co-expression with scsEst01; lanes 5–8: pKJE7 co-expression with scsEst01; lanes 9–12: pGTf2 co-expression with scsEst01; lanes 13–16: pGro7 co-expression with scsEst01; lanes 17–20: pG-KJE8 co-expression with scsEst01; lane M: unstained protein molecular-weight marker(Fermentas, Canada). Arrows: expected proteins. |
A 2.5-L-scale fermentation was performed using pET-28a-scsEst01 co-expressed with chaperone plasmid set pKJE7. The His-tag fusion protein in the soluble fraction was purified using an immobilized metal ion affinity column containing chelating Sepharose charged with Ni2+. The purified scsEst01 protein displayed a distinct b and on the SDS-PAGE gel(Fig. 4). The molecular weight of recombinant His-tagged scsEst01 was 35.5 kDa.
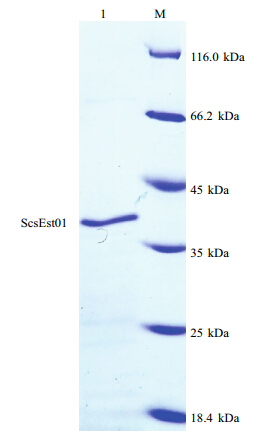 |
| Fig. 4 Sodium dodecyl sulfate-polyacrylamide gelelectrophoresis of purified recombinant His-tagged
scsEst01 Lane M: unstained protein molecular-weight marker(Fermentas, Canada); lane 1: purified scsEst01 protein, 35.5 kDa. |
As measured by Bradford’s method, the concentration of scsEst01 protein was 147.576 μg/ mL. The specific activity of purified scsEst01, determined using p-NP butyrate as a substrate, was found to be 171.03 U/mg under st and ard reaction conditions.
We measured the optimum activity of scsEst01 over a temperature range of 25–50°C and a pH range of 5.0–9.2. ScsEst01 showed the highest activity at 35°C(Fig. 5a) and pH 8.0(Fig. 5b), classifying it as a meso-thermophilic alkaline esterase. Thermostability analysis showed that scsEst01 was stable at -20 to 20°C and was inactivated at 40°C(Fig. 5c). The analysis of pH stability indicated that scsEst01 had a wide pH adaptation, being stable at pH values of 5.6– 8.6(Fig. 5d). ScsEst01 enzyme activity was strongly increased by Fe2+ and Mn2+, resulting in levels 1.5–2 times higher than in the absence of metal ions(Fig. 6a). The addition of 1% SDS inactivated scsEst01, while 1% Tween 80 and Tween 20 increased enzyme activity to 160% and 120%, respectively(Fig. 6b). In contrast, scsEst01 exhibited no tolerance to most organic solvents. Methanol, ethanol, isopropanol, acetone, DMSO, and DMF at final concentrations of 15% strongly inhibited scsEst01 catalytic activity(Fig. 6c).
 |
| Fig. 5 Effects of temperature and pH on scsEst01 activity Enzyme activity was measured under st and ard conditions. a. effect of temperature on scsEst01 enzyme activity. Activity was determined at pH 8.0 over 25–50°C with p-NP butyrate as the substrate. The value obtained at 35°C is shown as 100%; b. effect of pH on scsEst01 activity. ScsEst01 activity was measured over a pH range of 5.0–9.2 at 35°C with different pH buffers: 100 mmol/L citrate buffer(pH 5.0–6.5), 100 mmol/L potassium phosphate buffer(pH 6.5–7.5), 100 mmol/L Tris-HCl buffer(pH 7.5–9.0), and 50 mmol/L CHES buffer(pH 9.0–9.2); c. thermostability of scsEst01. The enzymatic assay was performed at 35°C following incubation of scsEst01 for 30 min at -20, 0, 10, 20, 30, 40, 50 and 60°C for 30 min. The value obtained at -20°C is shown as 100%; d. pH stability of scsEst01. The enzyme was mixed with equal volumes of various pH phosphate buffers(pH range of 5.0–9.2)at 4°C overnight. Enzyme activity was then tested at 35°C in 100 mmol/L Tris-HCl buffer(pH 8.0)with p-NP butyrate used as the substrate. The value obtained at pH 7.4 is shown as 100%. |
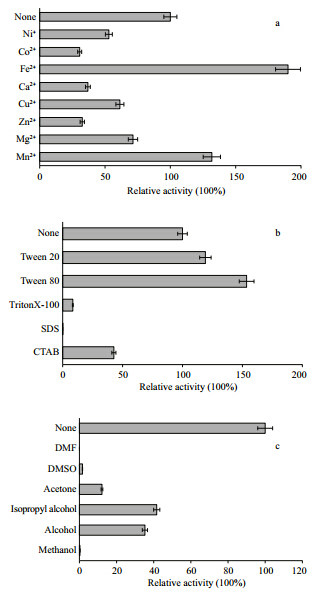 |
| Fig. 6 Effects of metal ions, detergents and organic solvents
on scsEst01 esterase activity The enzymatic assay was performed at 35°C in 100 mmol/L TrisHCl buffer(pH 8.0)with p-NP butyrate used as the substrate. Enzyme activity in the absence of additives was defined as 100%. a. effects of metal ions on scsEst01 esterase activity. The metal ions(Ni+, Co2+, Fe2+, Ca2+, Cu2+, Zn2+, Mg2+ and Mn2+)were added to solution to a final concentration of 10 mmol/L; b. effects of detergents on scsEst01 activity. Detergent effects were examined using 1% Tween 20, Tween 80, TritonX-100, SDS, and CTAB; c. effects of organic solvents on scsEst01 activity. Organic solvents were added to give a concentration of 15%. |
4 DISCUSSION The marine environment is diverse and contains a novel pool of enzymes. A metagenomic approach is useful for discovery of various enzymes from this environment. In our study, we first constructed a metagenomic library from South China Sea sediment to screen for lipolytic enzymes based on functional expression. Because no positive fosmid clones were obtained, we designed specific primers to amplify the esterase genes using metagenomic DNA as a template. As a result, the scsEst01 gene was cloned and annotated as an esterase gene. Several lipolytic enzymes from deep-sea sediment metagenomes have been previously reported, such as h1Lip1(Hardeman and Sjöling, 2007), EM2L8(Park et al., 2007), estAT1 and estAT11(Jeon et al., 2008), EML1(Jeon et al., 2009), FLS18(Hu et al., 2010) and est6(Jiang et al., 2012). In this study, the esterase scsEst01 shared less than 90% identity with other lipolytic enzymes in the nr database, suggesting that it might be a novel esterase.
On the basis of its placement in a phylogenetic tree of amino acid sequences, scsEst01 was assigned to lipolytic enzyme family IV(mammalian HSL). The consensus pentapeptide with the active-site serine residue, GDSAG, was predicted to be the catalytic triad. The HGGG motif may be the oxyanion hole. This family of lipolytic enzymes is very common in marine sediment environments, and the conserved sequence of this family was once thought to be linked to cold adaption. Esterases from psychrophilic, mesophilic and thermophilic microorganisms share common motifs with HSL, however, indicating that temperature adaptation is not responsible for such extensive sequence conservation(Arpigny and Jaeger, 1999; Hu et al., 2010).
Many genes are frequently expressed as inactive inclusion bodies in heterologous hosts such as E.coli cells, usually as a consequence of rare codon usage (Nakamura et al., 2000; Jia and Li, 2005). Various strategies can be used to promote protein refolding and form soluble protein, such as optimization of induction conditions including temperature, pH and IPTG concentration. In a previous study, a large quantity of soluble EM2L8 protein was obtained by lowering the induction temperature from 37°C to 18°C(Park et al., 2007). Another strategy is to use a different host strain; for example, E.coli Rosetta (DE3)or Transetta(DE3)might be used instead of E. coli BL21(DE3), because BL21(DE3)strains lack of rare codons. In our study, however, optimizing induction conditions and changing host strains did not alter the outcome, as ScsEst01 was still expressed as an inclusion body. We next turned to chaperones, which have been successfully used to recover active recombinant proteins(Thomas et al., 1997; Nishihara et al., 1998, 2000). Chaperone plasmid sets with pET- 28a-scsEst01 were co-transformed into E.coli BL21 (DE3). Use of the chaperone plasmid pKJE7 promoted the formation of soluble recombinant scsEst01. The pKJE7 plasmid encodes chaperones dnaK-dnaJ-grpE that promote soluble expression of scsEst01 proteins. Large amounts of soluble scsEst01 were obtained through this co-expression.
We hoped to obtain a highly active esterase for industrial applications. The purified esterase scsEst01 had an enzyme activity of 171.03 U/mg, which was higher than 150.2 U/mg of Lipo1(Roh and Villatte, 2008), 41.8 U/mg of est25(Kim et al., 2006), 31.1 U/ mg of EstCE1(Elend et al., 2007), 104.41 U/mg of est6(Jiang et al., 2012), 143 U/mg of EstA(Chu et al., 2008) and 156 U/mg of EM2L8(Park et al., 2007), but lower than 345.9 U/mg of FLS18(Hu et al., 2010), 513.6 U/mg of EstA3(Elend et al., 2007) and 981 U/ mg of EstB(Chu et al., 2008). Because ScsEst01 easily forms inclusion bodies, the potential exists to further increase its enzyme activity by minimizing inclusion body formation through optimization of various fermentation conditions.
Except in the vicinity of hydrothermal vents and oceanic volcanoes, temperatures of seafloor environments are low, 2–4°C. Lipolytic enzymes from deep-sea sediment might thus be assumed to be coldadapted. In fact, however, the optimum temperature for most lipolytic enzyme activity is in the mesophilic or thermophilic range, except for est6, which exhibits its highest activity at 20°C(Jiang et al., 2012), and similarly EML1 at 25°C(Jeon et al., 2009). The coldadapted esterase EM2L8 from a deep-sea sediment metagenome has an optimum temperature at 50–55°C (Park et al., 2007). The low-temperature active lipase hiLip1 shows optimal activity at 35°C(Hardeman and Sjöling, 2007), while the cold-adapted lipolytic enzymes FLS18 and EstF from the South China Sea deep-sea sediment metagenome have optimal activities at 50°C(Hu et al., 2010; Fu et al., 2011). All these results support the viewpoint that the optimal temperature of enzyme activity is usually higher than the optimal growth temperature of the enzyme hosts (Sheridan et al., 2000).
ScsEst01, an alkaline esterase, has optimal activity at 35°C and pH 8.0. Tween 20 and Tween 80 at appropriate concentrations increase scsEst01 activity. In addition, scsEst01 activity is strongly increased by Fe2+ to levels about two times higher than without metal ions. ScsEst01 is therefore a potential c and idate esterase for industrial applications.
5 CONCLUSIONTo summarize, the novel esterase scsEst01 was recombinantly expressed from a South China Sea sediment metagenome and assigned to lipolytic family IV. The optimum temperature for scsEst01 activity is higher than that of enzyme’s original cold environment. Characterization of scsEst01 indicated that the enzyme is an alkaline esterase that shows intolerance to organic solvents. Our study has demonstrated that deep-sea sediments are a very important source of novel lipolytic enzymes. Further overexpression, purification and biochemical characterization(such as substrate specificity)of scsEst01 is planned to assess its potential for industrial applications.
| Amann R I, Ludwig W, Schleifer K H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev., 59 (1): 143-169. |
| Arpigny J L, Jaeger K E. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J., 343 : 177-183. |
| Bayer S, Kunert A, Ballschmiter M, Greiner-Stoeffele T. 2010. Indication for a new lipolytic enzyme family: isolation and characterization of two esterases from a metagenomic library. J. Mol. Microbiol. Biotechnol., 18 (3): 181-187. |
| Bornscheuer U T. 2002. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev., 26 (1): 73-81. |
| Bradford M M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72 (1-2): 248-254. |
| Chu X M, He H Z, Guo C Q, Sun B L. 2008. Identification of two novel esterases from a marine metagenomic library derived from South China Sea. Appl. Microbiol. Biotechnol., 80 (4): 615-625. |
| Elend C, Schmeisser C, Hoebenreich H, Steele H L, Streit W R. 2007. Isolation and characterization of a metagenomederived and cold-active lipase with high stereospecificity for (R)-ibuprofen esters. J. Biotechnol., 130 (4): 370-377. |
| Fu C Z, Hu Y F, Xie F, Guo H, Ashforth E J, Polyak S W, Zhu B L, Zhang L X. 2011. Molecular cloning and characterization of a new cold-active esterase from a deep-sea metagenomic library. Appl. Microbiol. Biotechnol., 90 (3): 961-970. |
| Handelsman J, Rondon M R, Brady S F, Clardy J, Goodman R M. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol., 5 (10): R245-R249. |
| Hardeman F, Sjöling S. 2007. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol., 59 (2): 524-534. |
| Hu Y F, Fu C Z, Huang Y P, Yin Y S, Cheng G, Lei F, Lu N, Li J, Ashforth E J, Zhang L X, Zhu B L. 2010. Novel lipolytic genes from the microbial metagenomic library of the South China Sea marine sediment. FEMS Microbiol. Ecol., 72 (2): 228-237. |
| Jeon J H, Kim J T, Kang S G, Lee J H, Kim S G. 2008. Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar. Biotechnol., 11 (3): 307-316. |
| Jeon J H, Kim J T, Kim Y J, Kim H K, Lee H S, Kang S G, Kim S J, Lee J H. 2009. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol., 81 (5): 865-874. |
| Jia M W, Li Y D. 2005. The relationship among gene expression, folding free energy and codon usage bias in Escherichia coli. FEBS Lett., 579 (24): 5 333-5 337. |
| Jiang X W, Xu X W, Huo Y Y, Wu Y H, Zhu X F, Zhang X Q, Wu M. 2012. Identification and characterization of novel esterases from a deep-sea sediment metagenome. Arch. Microbiol., 194 (3): 207-214. |
| Kennedy J, Marchesi J R, Dobson A D W. 2008. Marine metagenomics: strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb.Cell Fact., 7 : 27. |
| Kim E Y, Oh K H, Lee M H, Kang C H, Oh T K, Yoon J H. 2009. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl. Environ. Microbiol., 75 (1): 257- 260. |
| Kim Y J, Choi G S, Kim S B, Yoon G S, Kim Y S, Ryu Y W. 2006. Screening and characterization of a novel esterase from a metagenomic library. Protein Expres. Purif., 45 (2): 315-323. |
| Lämmle K, Zipper H, Breuer M, Hauer B, Buta C, Brunner H, Rupp S. 2007. Identification of novel enzymes with different hydrolytic activities by metagenome expression cloning. J. Biotechnol., 127 (4): 575-592. |
| Lee M H, Hong K S, Malhotra S, Park J H, Hwang E C, Choi H K, Kim Y S, Tao W X, Lee S W. 2010. A new esterase EstD2 isolated from plant rhizosphere soil metagenome. Appl. Microbiol. Biotechnol., 88 (5): 1 125-1 134. |
| Lee M H, Lee C H, Oh T K, Song J K, Yoon J H. 2006. Isolation and characterization of a novel lipase from a metagenomic library of tidal flat sediments: evidence for a new family of bacteria lipases? Appl. Environ. Microbiol., 72 (11): 7 406-7 409. |
| Nakamura Y, Gojobori T, Ikemura T. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucl. Acids Res., 28 (1): 292. |
| Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. 1998. Chaperone coexpression plasmids: Differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escheric h ia coli. Appl. Environ. Microbiol., 64 (5): 1 694-1 699. |
| Nishihara K, Kanemori M, Yanagi H, Yura T. 2000. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol., 66 (3): 884-889. |
| Park H J, Jeon J H, Kang S G, Lee J H, Lee S A, Kim H K. 2007. Functional expression and refolding of new alkaline esterase, EM2L8 from deep-sea sediment metagenome. Protein Expres. Purif., 52 (2): 340-347. |
| Ranjan R, Grover A, Kapardar R K, Sharma R. 2005. Isolation of novel lipolytic genes from uncultured bacteria of pond water. Biochem. Biophys. Res. Commun., 335 (1): 57-65. |
| Roh C, Villatte F. 2008. Isolation of a low-temperature adapted lipolytic enzyme from uncultivated micro-organism. J. Appl. Microbiol., 105 (1): 116-123. |
| Schmeisser C, Steele H, Streit W R. 2007. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol., 75 (5): 955-962. |
| Sheridan P P, Panasik N, Coombs J M, Brenchley J E. 2000. Approaches for deciphering the structural basis of low temperature enzyme activity. Biochim. Biophys. Acta, 1543 (2): 417-433. |
| Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol., 24 (8): 1 596-1 599. |
| Thomas J G, Ayling A, Baneyx F. 1997. Molecular chaperones, folding catalysts, and the recovery of active recombinant proteins from E. coli. To fold or to refold. Appl. Biochem. Biotechnol., 66 (3): 197-238. |
| Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res., 25 (24): 4 876- 4 882. |
 2015, Vol. 33
2015, Vol. 33


