Shanghai University
Article Information
- LI Yanyu (李艳宇), ZHANG Tao (张涛), ZHANG Congyao (张丛尧), ZHU Ying (朱莹), DING Jianfeng (丁鉴锋), MA Yuexin (马悦欣)
- Bacterial diversity in the intestine of young farmed puffer fish Takifugu rubripes
- Chinese Journal of Oceanology and Limnology, 2015, 33(4): 913-918
- http://dx.doi.org/10.1007/s00343-015-4219-2
Article History
- Received Sep. 3, 2014;
- accepted in principle Oct. 31, 2014;
- accepted for publication Jan. 20, 2015
2 Dalian Tianzheng Industry Co. Ltd., Dalian 116011, China
Puffer fish(Takifugu rubripes is one of the most important farmed fish species in China,Japan and Korea. In 2007,annual production in China was approximately 8 000 metric tonnes with a value of 600 million Yuan RMB(Gao et al., 2011). In recent years,yields have varied owing to fluctuations in dem and for the export trade. The interaction between gut microbiota and innate immunity in fish has been reviewed by Gómez and Balcázar(2008),who concluded that normal microbiota confers numerous benefits on the intestinal physiology of the host. Some of these benefits include the metabolism of nutrients and organic substrates substances, and their contribution to the phenomenon of colonization resistance. The intestinal microflora of T. rubripes has been analyzed via culture methods and clone library screening(Sugita et al., 1988,2010; Chen et al., 2013). Tanasomwang and Muroga(1989)studied the microbiota associated with the larvae and juveniles of T. rubripes and other species. Kim et al.(2007)found that the bacterial diversity of the intestinal contents differed from that of the gut mucus in rainbow trout(Oncorhynchus mykiss). The aims of this study were to identify the dominant bacterial populations associated with the intestinal mucus of farmed puffer fish via 16S rRNA gene sequencing of DNA from bacterial isolates, and to compare the microflora sampled at 2-month intervals. 2 MATERIAL AND METHOD 2.1 Sampling of puffer fish
Puffer fish(T. rubripes and )were reared in a 64-m3 tank and fed a natural diet(fresh fish,mainly Ammodytespersonatus)twice a day(7:00 and 17:00)at a rate of 2% body weight. Water in the tank was replaced daily with fresh underground brine(salinity 27(Practical Salinity Units),pH 7.5)adjusted to ~20°C via a heat exchanger. Low pressure electric air pumps provided aeration via air stones,maintaining dissolved oxygen levels at 6.5 mg/L. Water temperature was maintained at 20–22°C. After a >4- and >6-month rearing period,young fish(weight ~50 g,n=3 and weight ~110 g,n=2,respectively)were collected from Dalian Tianzheng Industry Co. Ltd.,Dalian,China,in September and November, and transported alive to the laboratory at Dalian Ocean University. Specimens were held in plastic tanks(200 L)at a water temperature of 19–20°C for 2 days and deprived of food to empty the intestine. Low pressure electrical air pumps provided aeration via air stones. Salinity ranged from 32 to 33 and pH ranged 7.9–8.1. 2.2 Isolation of culturable intestinal microbiota
Fish were sacrificed and their external surfaces sterilized using 70% ethanol(Luczkovich and Stellwag, 1993). The entire intestine was removed by aseptic surgery and the intestines were weighed,pooled, and homogenized in sterile physiological saline using a manual glass homogenizer. Appropriate dilutions were prepared and 0.01-mL aliquots were spread over the surface of agar plates of ZoBell 2216E medium and incubated at 25°C for 7 days. A total of 60 colonies were picked at r and om from the plates. Pure cultures were obtained by repeated subculture on new agar plates and stored at -80 °C as suspensions in Zobell broth with 30%(v/v)glycerol(Liu et al., 2012). 2.3 Sequencing of the 16S rRNA gene
Bacterial cells were grown on ZoBell 2216E and harvested in the late exponential phase of growth. Genomic DNA was extracted as previously described(Wu et al., 2010). A partial 16S rRNA gene of each isolate was amplified using the F27/R1492 primer set(Lane,1991)in 50 μL of reaction mixture,which contained 5-μL 10×PCR buffer,4-μL template,250-μmol/L dNTP,1.5-mmol/L MgCl 2,0.4-μmol/L of each primer and 0.25 U Taqpolymerase. Conditions were as follow: initial denaturation step of 4 min at 94°C,followed by 30 cycles of a 30-s denaturation at 94°C,30 s of annealing at 56 °C, and a 30-s extension at 72°C,followed by a final extension step of 10 min at 72°C. Amplification products were analyzed by electrophoresis in 1%(w/v)agarose gel containing GoldView. Sequencing reactions of purified PCR products were performed using an ABI 3730KL DNA sequencing system. 2.4 Analysis of 16S rRNA gene sequences
Sequences of all isolates were submitted for similarity searches with the RDP SEQMATCH program(http://rdp.cme.msu.edu/)after unreliable sequences at the 3′- and 5′-ends were removed. For phylogenetic analyses,the sequences were aligned using the ClustalW program. The construction of a neighbor-joining tree was based on Kimura-2-parameter distances, and bootstrap analysis of 1 000 re-samplings was performed using MEGA 4(Tamura et al., 2007).
To compare bacterial diversity between the two sampling dates,Shannon’s index was calculated to reflect species richness of the bacterial community(Shannon,1948). Species evenness of the community was estimated with Pielou’s diversity(Pielou,1966). Bray-Curtis index(Bray and Curtis, 1957) and Sørensen index(Sørensen,1948),which reveal similarities between communities,were also computed. 3 RESULT
Initially,60 nucleotide sequences from 16S rRNA genes were obtained from the cultured microflora in the intestine of young farmed puffer fish. Using 99% minimum similarity as the threshold for any pair of sequences in a phylotype(Kim et al., 2007),33 bacterial phylotypes were retrieved. The phylogenetic tree(Fig. 1)shows the closeness of the evolutionary relationships of the 33 representative isolates. The closest strains were determined by the RDP SEQMATCH program, and the results displayed in Table 1.
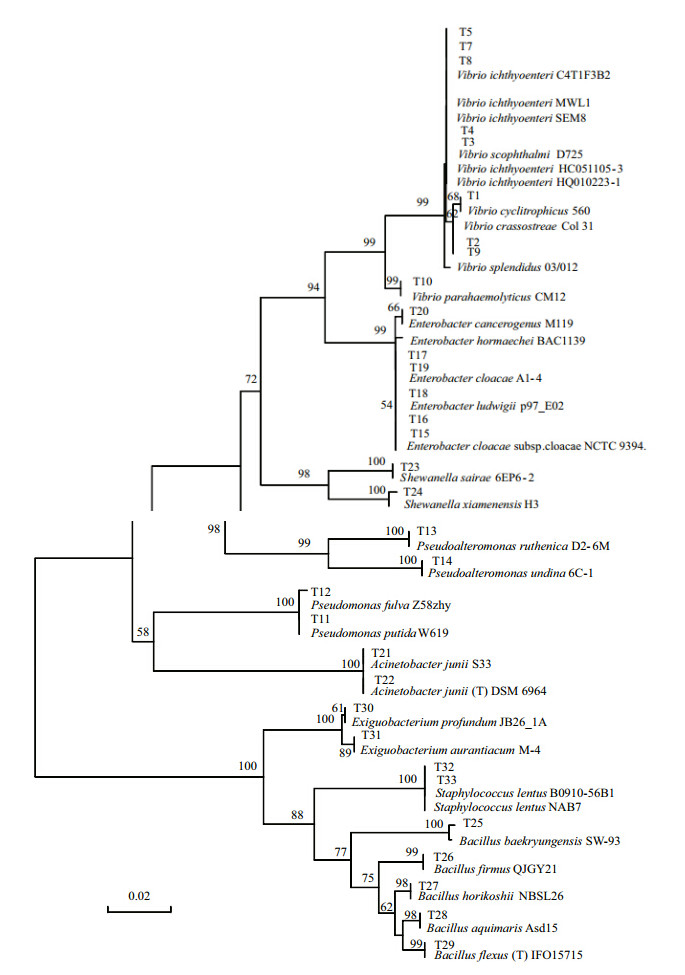 |
| Fig. 1 Neighbor-joining phylogenetic tree showing the relationships of 33 16S rRNA gene sequences retrieved from the intestines of young farmed puffer fish
Bootstrap values,based on 1 000 re-samplings,display the significance of the interior nodes and are shown at branch points; only values displaying >50% are given. The scale bar represents a 2% estimated difference in nucleotide sequences. |
 |
All isolates were assigned to Proteobacteria and Firmicutes,with the dominant Phylum of culturable intestinal microbiota being Proteobacteria(68.3%). At the genus level(Fig. 2),nine taxonomic groups were isolated from the intestine,with representatives of the genera Vibrio(16/60),Enterobacter(16/60) and Bacillus(14/60)accounting for 76.7% of the total.
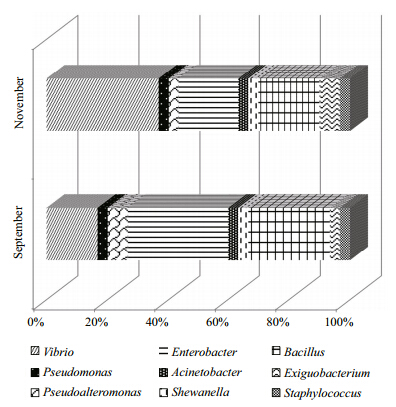 |
| Fig. 2 Culturable intestinal microbiota of puffer fish sampled in September and November(at genus level) |
The Shannon diversity index for the bacterial community was higher in November(2.83)than in September(2.68). The Pielou index in September and November was 0.91 and 0.94,respectively,indicating that species were evenly distributed at both sampling dates. The Bray-Curtis and Sorensen similarity indices(0.33 and 0.31,respectively)revealed that the bacterial communities differed completely between the two sampling dates.4 DISCUSSION
In agreement with previous studies(Sugita et al., 1988,2010; Chen et al., 2013),it was demonstrated that the class Gammaproteobacteria were predominantly isolates from puffer fish intestine. However,some culturable genera retrieved in this study,i.e.,Enterobacter,Bacillus,Exiguobacterium and Staphylococcus,have not been previously reported as part of the intestinal bacterial microflora in T. rubripes. Shewanella,Vibrio,Pseudoalteromonas,Pseudomonas and Acinetobacter were previously isolated from the intestinal content of T. rubripes(Sugita et al., 1988,2010; Chen et al., 2013). The disparity between our results and previous studies could be explained by the above-mentioned findings in rainbow trout that the bacterial diversity of gut mucus differed from that of the intestinal contents(Kim et al., 2007). The microbiota associated with the larval and juvenile stages of T. rubripes was characterized by predominating groups of Vibrio,Acinetobacter and Pseudomonas(Tanasomwang and Muroga, 1989). A number of bacterial genera were also recorded in other species of the same genus of puffer fish. For example,Vibrio,Pseudomonas,Acinetobacter and Staphylococcus were identified in the intestinal contents of Fugu(Takifugu)vermicularisvermicularis(Noguchi et al., 1987; Sugita et al., 1987) and F. niphobles(Sugita et al., 1987,1989). Vibrio,Bacillus,Pseudoalteromonas and Shewanellawere found in the intestine of T. obscurus(Yang et al., 2007).
Probiotic applications of Bacillus,Pseudomonas,Enterobacter and Vibrioin salmonids resulted in elevated health status, and improved disease resistance,growth performance,feed utilization,body composition,gastrointestinal colonisation and ,subsequently,microbial modulation(review by Merrifield et al., 2010; Burbank et al., 2011). Shewanellahas also been evaluated as a probiotic in the culture of gilthead seabream and Senegalese sole(reviewed by Tapia-Paniagua et al., 2012). However,whether the genera of bacteria found in this study have a probiotic effect in puffer fish culture is unclear and requires investigation.
Conversely, Vibrio ichthyoenteri from fin ulceration and liver tissue has been reported to be a pathogen of T. rubripes,while Bacillussp. RG-4BB from the intestine of F. rubripeswas demonstrated to produce tetrodotoxin(Wu et al., 2005). Regarding the isolates in the present study,an assessment of the virulence of V. ichthyoenteri in the puffer fish and the ability of Bacillusspp. to produce tetrodotoxin is warranted.
In the present study,five sequences retrieved via denaturing gradient gel electrophoresis(DGGE; data not shown)were related to an uncultured spirochete(AB194658,93%),Pseudomonas sp.(KF870448,94%),an uncultured bacterium(EU652651,96%),Arcobacter sp.(FN397894,94%) and Sphingobiumsp.(KJ372001,98%). Using DGGE,Yang et al.(2007)detected an uncultured spirochete sequence(AB194658)from the intestine of T. obscurus. Uncultured spirochetes(AB194657,AB194658)with length of 2.5–4.5 μm were observed microscopically in the intestinal contents of T. niphoblesat high densities and clone library screened(Shiina et al., 2006).
In this study,puffer fish sampled in September and November harbored different intestinal bacterial communities,which could be explained by variations in diet and /or changes in the intestinal microenvironment. 5 CONCLUSION
Vibrio,Bacillus and Enterobacter comprised the dominant culturable microbiota associated with the intestinal mucus of farmed puffer fish. This is the first record of Enterobacter,Bacillus,Exiguobacterium and Staphylococcus in the intestine of young T. rubripes from a commercial fish-farm.
| Bray J R, Curtis J T. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr., 27 (4): 325-349. |
| Burbank D R, Shah D H, LaPatra S E, Fornshell G, Cain K D. 2011. Enhanced resistance to coldwater disease following feeding of probiotic bacterial strains to rainbow trout (Oncorhynchus mykiss). Aquaculture, 321 (3-4): 185-190. |
| Chen C H, Tanaka Y, Komiya Y, Itoi S, Sugita H. 2013. Predominant intestinal bacteria of the pufferfish (Takifugu rubripes Temminck & Schlegel, 1850) reared in an indoor tank as determined by the clone library analysis and culture method. J. Appl. Ichthyol., 29 (6): 1 374-1 377. |
| Gao L J, Huang Y Q, Xia L J, Lu J X, Liu S C. 2011. Comparison of flesh quality of farmed fugu, Takifugu rubripes from different culture models. J. F ish. C hina, 35 (11): 1668- 1676. (in Chinese with English abstract) |
| Gómez G D, Balcázar J L. 2008. A review on the interactions between gut microbiota and innate immunity of fish. FEMS Immunol. Med. Microbiol., 52 (2): 145-154. |
| Kim D H, Brunt J, Austin B. 2007. Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhynchus mykiss). J. Appl. Microbiol., 102 (6): 1 654-1 664. |
| Lane D J. 1991. 16S/23S rRNA sequencing. In : Stackebrandt E, Goodfellow M D eds. Nucleic Acid Techniques in Bacterial Systematic. 1stedn. John Wiley and Sons, Chichester. p.115-175. |
| Liu Z M, Ma Y X, Yang Z P, Li M, Liu J, Bao P Y. 2012. Immune responses and disease resistance of the juvenile sea cucumber Apostichopus japonicus induced by Metschnikowia sp. C14. Aquaculture, 368 -369 : 10-18. |
| Luczkovich J J, Stellwag E J. 1993. Isolation of cellulolytic microbes from the intestinal tract of the pinfish, Lagodon rhomboides : size-related changes in diet and microbial abundance. Mar. Biol., 116 (3): 381-388. |
| Merrifield D L, Dimitroglou A, Foey A, Davies S J, Baker R T M, Bøgwald J, Castex M, Ringø E. 2010. The current status and future focus of probiotic and prebiotic applications for salmonids. Aquaculture, 302 (1-2): 1-18. |
| Noguchi T, Hwang D F, Arakawa O, Sugita H, Deguchi Y, Shida Y, Hashimoto K. 1987. Vibrio alginolyti c us, a tetrodotoxin-producing bacterium, in the intestines of the fish Fugu vermicularis vermicularis. Mar. Biol., 94 (4): 625-630. |
| Pielou E C. 1966. The measurement of diversity in different types of biological collections. J. T heor. B iol., 13 : 131-144. |
| Shannon C E. 1948. A mathematical theory of communication. The Bell Syst em Techn ical J ournal, 27 (3): 379-423. |
| Shiina A, Itoi S, Washio S, Sugita H. 2006. Molecular identification of intestinal microflora in Takifugu niphobles. Comp. Biochem. Physiol. Part D, 1 (1): 128- 132. |
| Sørensen T. 1948. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content, and its application to analyses of the vegetation on Danish commons. Kongelige Danske Videnskabernes Selskabs Biologiske Skrifter, 5 (4): 1-34. |
| Sugita H, Iwata J, Miyajima C, Kubo T, Noguchi T, Hashimoto K, Deguchi Y. 1989. Changes in microflora of a puffer fish Fugu niphobles, with different water temperatures. Mar. Biol., 101 (3): 299-304. |
| Sugita H, Noguchi T, Furuta M, Harada T, Murata O, Hashimoto K, Deguchi Y. 1988. The intestinal microflora of cultured specimens of a puffer Fugu rubripes rubripes. Nippon Suis an Gakkaishi, 54 (4): 733. |
| Sugita H, Noguchi T, Hwang D F, Furuta M, Motokane T, Sonoda T, Hashimoto K, Deguchi Y. 1987. Intestinal microflora of coastal puffer fishes. Nippon Suisan Gakkaishi, 53 (12): 2 201-2 207. |
| Sugita H, Sugiyama K, Itoi S. 2010. Culturable bacterial flora in the intestinal tract of Japanese pufferfish Takifugu rubripes. .Aquacult. Sci, 58 (3): 437-438. |
| Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol., 24 (8): 1 596-1 599. |
| Tanasomwang V, Muroga K. 1989. Intestinal microflora of rockfish Sebastes schlegeli, tiger puffer Takifugu rubripes and red grouper Epinephelus akaara at their larval and juvenile stages. Nippon Suisan Gakkaishi, 55 (8): 1 371- 1 377. |
| Tapia-Paniagua S T, Díaz-Rosales P, León-Rubio J M, de La Banda I G, Lobo C, Alarcón F J, Chabrillón M, Rosas-Ledesma P, Varela J L, Ruiz-Jarabo I, Arijo S, Esteban M A, Martínez-Manzanares E, Mancera J M, Balebona M C, Moriñigo M A. 2012. Use of the probiotic Shewanella putrefaciens Pdp11 on the culture of Senegalese sole (Solea senegalensis, Kaup 1858) and gilthead seabream (Sparus aurata L.). Aquacult. Int., 20 (6): 1 025-1 039. |
| Wu S G, Gao T H, Zheng Y Z, Wang W W, Cheng Y Y, Wang G T. 2010. Microbial diversity of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco). Aquaculture, 303 (1-4): 1-7. |
| Wu Z L, Yang Y, Xie L P, Xia G L, Hu J C, Wang S J, Zhang R Q. 2005. Toxicity and distribution of tetrodotoxinproducing bacteria in puffer fish Fugu rubripes collected from the Bohai Sea of China. Toxicon, 46 (4): 471-476. |
| Yang G M, Bao B L, Peatman E, Li H R, Huang L B, Ren D M. 2007. Analysis of the composition of the bacterial community in puffer fish Takifugu obscurus. Aquaculture, 262 (2-4): 183-191. |
 2015, Vol. 33
2015, Vol. 33


