Shanghai University
Article Information
- HUANG Wen (黄雯), XU Fei (许飞) , QU Tao (曲涛), LI Li (李莉), QUE Huayong (阙华勇), ZHANG Guofan (张国范)
- Iodothyronine deiodinase gene analysis of the Pacific oyster Crassostrea gigas reveals possible conservation of thyroid hormone feedback regulation mechanism in mollusks
- Chinese Journal of Oceanology and Limnology, 2015, 33(4): 997-1006
- http://dx.doi.org/10.1007/s00343-015-4300-x
Article History
- Received Nov. 6, 2014;
- accepted in principle Dec. 5, 2014;
- accepted for publication Jan. 4, 2015
2 University of Chinese Academy of Sciences, Beijing 100049, China
Thyroid hormones(THs),contain precursor L-thyroxine(T4) and active form 3,3′,5-triiodothyronine(T3), and play important roles in growth,development, and metabolism in vertebrates(Yen,2001). THs have been regarded as vertebrate-exclusive hormones in the past century. In the following years,successively reported evidences from physiological and molecular level strongly supported that TH related function was also presented in cephalochordate and urochordata(Laudet,2011). Thus,the origin and evolution of THs were dated back to chordates. Nevertheless,TH related studies in non-chordate invertebrates were scarce and scattered in several species: settlement and metamorphosis are induced by T4 and T3 in the two abalone species Haliotis discus and H. gigantean,respectively(Fukazawa et al., 2001); growth is promoted by T4 in the freshwater prawn Macrobrachium rosenbergii(Roustaian and Gaik, 2006); both growth and metamorphosis are promoted by T4 in the sea urchin Lytechinus variegates(Heyland et al., 2006); one thyroid hormone receptor(TR) and one iodothyronine deiodinase homolog have been identified in thetrematode Schistosoma mansoni and the scallop Chlamys farreri,respectively(Wu et al., 2007,2012). These physiological and molecular evidences indicated that THs should also be involved in certain biological processes, and the molecular mechanisms of TH metabolism might also exist in a wide range of non-chordate invertebrates.
As a key enzyme in the regulation of TH levels,iodothyronine deiodinase catalyzes the initiation and termination of TH effects, and could evidence the existence of TH metabolism. Deiodinases are classified into three types in vertebrates. Type II deiodinase(D2)catalyzes the outer ring deiodination(ORD)to transform prohormone T4 to active T3; type III deiodinase(D)catalyzes inner ring deiodination(IRD)to inactivate T4 and T3; Type I deiodinase(D1)is capable of both ORD and IRD(Bianco et al., 2002). Deiodinases are transmembrane enzymes with similar structural organizations; they belong to the thioredoxin(TRX)fold superfamily. Deiodinases contain a conserved active center with a rare amino acid,selenocysteine(SeC),encoded by an in-frame TGA codon. Those proteins with SeC incorporation are referred to as selenoproteins. The TGA codon is recognized as a stop codon in the vast majority of mRNAs. However,in deiodinase mRNA,a stem-loop structure called SeC insertion sequence(SECIS)element in the 3′untranslated region(3′-UTR)is necessary and sufficient to translate TGA as SeC instead of a stop codon(Köhrle,2002). To date,only one deiodinase has been identified in Urochordata from the ascidian Halocynthia roretzi(Shepherdley et al., 2003) and one in non-chordate invertebrates from the scallop Chlamys farreri(Wu et al., 2012). As a result,the deiodinase genes,as well as their related hormone signaling network,are largely unknown in invertebrates. Identifying deiodinase orthologs in major invertebrate clades is thus necessary to reveal the origin and evolution of thyroid hormone regulation systems in animals.
As a well-known species in mollusks,the Pacific oyster Crassostrea gigas(Thunberg)has received much attention for its importance in aquaculture as well as in ecosystems and evolutionary studies. The genome of the Pacific oyster was released in 2012(Zhang et al., 2012), and several pivotal orthologs in the TH signaling pathway have been identified,including two iodothyronine deiodinases,one TR, and some thyroid peroxidases. In this study,we cloned the cDNAs and promoters of two deiodinases in C. gigas. Furthermore,one deiodinase in the mollusk Lottia gigantea and six in the annelid Capitella teletawere found by tblastn searches using available genome data. A phylogenetic analysis was conducted to investigate the evolution of these iodothyronine deiodinases. Then,the spatiotemporal expression patterns of two oyster deiodinases were analyzed to gain insight into their function. Finally,T4 treatment and Epinephrine(EPI)stimulation experiments were conducted to investigate the regulation of oyster deiodinase synthesis. 2 MATERIAL AND METHOD 2.1 Treatment and sample collection
The Pacific oysters used in this study were collected from a cultured population in Qingdao,China. Larval development was monitored by light microscopy. The sampled developmental stages included the egg(E),blastula(B),gastrula(G),trochophore(T1 and T2),D-shaped larvae(D1,D2, and D3),umbo larvae(U1,U2, and U3),pediveliger larvae(P),spat,(S) and juvenile(J)stages. Adult organ samples,including samples of the labial palps,gills,hemolymph,gonad,digestive gl and ,mantle, and adductor muscle were collected from three individual oysters. Pediveliger larvae were treated with T4 and EPI. Primary T4(Sigma,St. Louis,MO,USA)stock was prepared at a concentration of 1.29×10-4mol/L in 0.01 mol/L NaOH solution. The EPI(Sigma)stock was prepared at a concentration of 0.1 mol/L. T4 treatment was conducted by continuous immersion of pediveliger larvae in sea water with 1.29×10-8mol/L T4,while EPI stimulation was performed by short-term immersion of pediveliger larvae in sea water with 10-4mol/L EPI for 20 min. Larvae were then raised in filtered sea water. Pediveliger larvae were sampled at 3 h,6 h,12 h,24 h, and 48 h post T4 treatment and 20 min,1 h,3 h,6 h,12 h, and 24 h after EPI stimulation. All samples were frozen in liquid nitrogen, and then transferred to a -80°C refrigerator for long-term storage. 2.2 RNA isolation and cDNA synthesis
Total RNA was extracted using TRIzol reagent(Invitrogen,Carlsbad,CA,USA)according to the manufacturer’s instructions. The concentration of each RNA sample was measured using a NanoDrop 2000 spectrophotometer(Thermo Scientific,Waltham,MA,USA). The integrity of the RNA samples was assessed by agarose gel electrophoresis. First-str and cDNA was with gDNA Eraser(TaKaRa,Shiga,Japan)following the manufacturer’s instructions. 2.3 Cloning of oyster deiodinase cDNAs and promoters
Primers(CgDx-F,CgDx-R,CgDy-F, and CgDy-R; Table 1)were designed according to the incomplete coding sequences(CDS)of oyster deiodinases to amplify the middle fragment. The PCR products were cloned into pMD19-T simple vectors(TaKaRa) and sequenced by Sangon Biotech(Shanghai,China). Based on the sequencing results,gene-specific primers(listed in Table 1)were designed for the 3′ and 5′ends for rapid amplification of cDNA ends(RACE),which was conducted as previously described(Qu et al., 2014). The oyster deiodinases(designated as CgDx and CgDy)were located in the Pacific oyster genome(Zhang et al., 2012). The genomic regions ~3 kb upstream of CgDx and CgDywere used as references to design promoter-specific forward primers, and the reverse primers were designed based on the 5′-UTR sequence.
Quantitative real-time PCR(qRT-PCR)was performed in a 96-well plate using the ABI 7500 FAST Real-Time PCR System(Foster City,CA,USA). Two CgDx-specific primers(qCgDx-F and qCgDx-R; Table 1) and two CgDy-specific primers(qCgDy-F and qCgDy-R; Table 1)were used. Each PCR reaction mixture included 2 μL of cDNA template,10 μL of 2× SYBR ®Green PCR Master Mix(ABI),0.4 μmol/L each of the forward and reverse primers, and sterile water to a 20 μL final volume. The amplification program was as follows: 50°C for 2 min,95°C for 2 min,followed by 40 cycles of 95°C for 3 s, and 60°C for 30 s. Melt curves were generated at the end of the cycling stage to confirm the specificity of the primers. For the various developmental stages and the T4 and EPI treatments,the ribosomal protein S18( RS18)gene was used as an internal control(Du et al., 2013), and for the different organs,the elongation factor(EF)gene was used as previously reported(Zhang et al., 2011). The threshold cycle(Ct)was automatically calculated using 7500 Software v2.0.1(ABI). The 2-ΔΔCtmethod was used to calculate the expression level of target genes. 2.5 Database searching
A homology search for non-chordate invertebrate deiodinases was conducted using tblastn against the genome database at the National Center for Biotechnology Information(http://www.ncbi.nlm.nih.gov/)or the Joint Genome Institute(http://genome.jgi.doe.gov/). The amino acid sequences of CgDx and CgDy were used as queries. Any sequence with conserved amino acids surrounding SeC(RPLXXXFGSCTUPPF,with two mismatches allowed)were retained. Then,an ~3 kb sequence was extracted from the upstream and downstream regions of the retained promoter sequences. These sequences were submitted to ORF Finder(http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and checked manually to attain whole amino acid sequences. The thyroid hormone responsive element(TRE)was identified by searching the promoter sequence and its complementary str and with the fuzznuc program in the EMBOSS suite of programs(http://genome.csdb.cn/cgi-bin/emboss/fuzznuc). The DNA element containing two direct repeats of variable half-sites(A/G)(A/G)GT(A/C/T)A spaced with 0–6 nucleic acids(designated as DR0–DR6)was used as a query(Das et al., 2009). 2.6 Phylogenetic tree construction
The phylogenetic tree was constructed from the deduced amino acid sequences of invertebrate deiodinases and representative vertebrate deiodinases. Amino acid sequences of deiodinases were aligned with ClustalW and a phylogenetic analysis of the data was carried out using the Maximum Likelihood algorithm implemented in MEGA5.0(http://www.megasoftware.net). The confidence values for each node of the inferred phylogeny were obtained by bootstrapping with 100 replicates. 2.7 Statistical analysis
All experiments were performed in triplicate. The comparison of gene expression levels were performed using the Student’s ttest. P<0.05 were considered statistically significant. 3 RESULT 3.1 Cloning of the full-length oyster deiodinase cDNAs and promoters
The full-length cDNAs of oyster iodothyronine deiodinases were cloned from C. gigas and were submitted to NCBI(accession numbers,CgDx: KM454867,CgDy: KM454868). The cDNA of CgDxcontained 1 645 nucleotides and the CDS contained 765 nucleotides,coding for a protein of 254 amino acids. The cDNA of CgDycontained 2 087 nucleotides and the CDS contained 735 nucleotides,coding for a protein of 244 amino acids. The CgDx and CgDycDNA sequences contained an in-frame TGA stop codon. The SeC insertion sequence elements(SECIS)in the 3′-UTRs of CgDx and CgDy(Fig. 1)were determined by the online tool SECISearch3(http://gladyshevlab.org/SelenoproteinPredictionServer/)(Mariotti et al., 2013),with the conserved nucleotides AUGA at the 5′base of the stem,AAA in the hairpin loop, and GA at the 3′base of the stem(Berry et al., 1993).
 |
| Fig. 1 The SECIS elements predicted by SECISearch3(Mariotti et al., 2013)in the CgDx(a) and CgDy(b)3′-UTRs
The nucleotides forming the SECIS core are marked in gray and the conserved unpaired nucleotides in the apical loop are also marked in gray and circled. Moreover,paired nucleotides are marked in light gray. The Infernal and Covels scores,free energy of the structure,position, and SECIS grade are listed. |
To investigate the regulation of oyster deiodinase synthesis,~3 kb of the promoter sequences of CgDx and CgDywere cloned, and one putative TRE(DR6)in the CgDxpromoter and two putative TREs(DR1)in the CgDypromoter were found. The localization and sequence details are listed in Fig. 2.
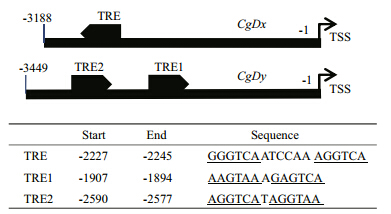 |
| Fig. 2 Schematic of the putative TREs in the CgDx and CgDy promoter regions
Arrows indicate the orientation of each TRE. The sequences and the positions(relative to TSS)of the putative TREs are listed. Conserved half sites are underlined. TRE,thyroid hormone responsive element; TSS,transcriptional start sites. |
Potential deiodinases were identified by tblastn, and one deiodinase from the mollusk Lottia gigantea(designated as LgDx) and six from the annelid Capitella teleta(designated as CtDato CtDf)were found. The details of those putative deiodinases are listed in Table 2.
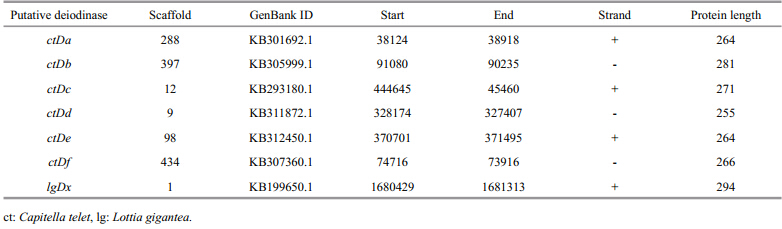 |
The alignment(Fig. 3)exhibited a putative transmembrane domain segment and three conserved secondary structures. The thioredoxin-fold βαβmotif was the most highly conserved,with a conserved region about 15 amino acids(RPLXXXFGSC(T/S)UP(P/S)F)surrounding the essential SeC(U)residue. The residues essential for deiodinase activity in C. gigaswere identical in structure to mouse D3(Schweizer et al., 2014).
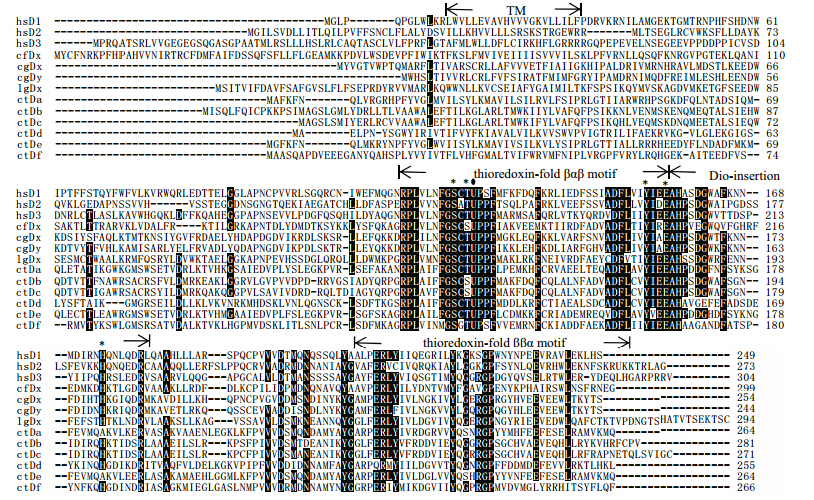 |
| Fig. 3 Sequence alignment of non-chordate invertebrate deiodinases with the three human deiodinases
The position of the putative transmembrane domain(TM)segment and the conserved secondary structures of deiodinases are shownabove the alignment. The essential SeC(U)are indicated with black circles above the alignment and the amino acids essential for deiodinase activity reported in Schweizer et al.(2014)are indicated by black asterisks. ctDa-f and lgDx are deiodinase homologs,which were found in genomes of Capitella telet and Lottia gigantean. The genome details of them were listed in Table 2. Human: Homo sapiens(hsD1: NP_000783.2; hsD2: NP_000784.2; hsD3: NP_001353.4); scallop: Chlamysfarreri(cfDx: AEX08671.1); ct: Capitella telet; lg: Lottia gigantean; cg: Crassostrea gigas. |
In the phylogenetic tree(Fig. 4),all invertebrate deiodinases were clustered with the vertebrate deiodinases subgroup and separated from the glutathione peroxidases(GPx),members of thioredoxin like superfamily served as out groups. This result indicated that deiodinases originated from a common ancestor gene. Vertebrate deiodinases were clustered together and formed a subgroup,indicating that gene duplication occurred in the common ancestor of vertebrates and subsequently differentiated to specific catalytic functions. With high similarity of 64.57%,CgDx and CgDy clustered outside of the vertebrate subgroups,suggesting that they underwent duplication independently. The six putative Capitella teletadeiodinases also clustered together.
 |
| Fig. 4 Phylogenetic tree of the known deduced amino acid sequences of the vertebrate iodothyronine deiodinases and the invertebrate deiodinase homologs
The phylogenetic tree was constructed by the Maximum Likelihood algorithm using MEGA5.0 software. CgDx and CgDyare marked with black triangles. ctDa-f and lgDx are deiodinase homologs,which were found in genomes of Capitella telet and Lottia gigantean. The genome details of them were listed in Table 2. hsGPx and mmGPx,members of thioredoxin like superfamily,are used as out groups. Human: Homo sapiens(hsD1: NP_000783.2; hsD2: NP_000784.2; hsD3: NP_001353.4; hsGPx: CAA41228); mouse: Mus musculus(mmD1: NP_031886.3; mmD2: NP_034180.1; mmD3: NP_742117.2; mmGPx: CAA27558); african clawed frog: Xenopus laevis(xlD1: NP_001089136.1; xlD3: NP_001081332.1); western clawed frog: Xenopus tropicalis(xtD2: NP_001184161.2); zebrafish: Danio rerio(drD1: NP_001007284.2; drD2: NP_997954.2; drD3: NP_001171406.2); atlantic halibut: Hippoglossus hippoglossus(hhD1: ABI93488.2; hhD2: ABI93490.2; hhD3: ABI93489.2); ascidians: Halocynthiaroretzi(hrDx: AAR25890.1); scallop: Chlamys farreri(cfDx: AEX08671.1); ct: Capitella telet; lg: Lottia gigantean; cg: Crassostrea gigas. |
qRT-PCR was performed to determine the expression patterns of oyster deiodinases in developmental stages and different organs. As shown in Figs.5 and 6,CgDx and CgDyhad distinct expression profiles,which indicated functional differentiation of the two oyster deiodinases.
Maternal expression of CgDx in the eggs was observed and the expression peaked at the blastula(B) and juvenile(J)stages separately; expression in both stages differed significantly from that in the egg(P<0.05). The expression level at the other stages was not significantly different from that of egg(Fig. 5a). However,the CgDytranscripts were undetectable in eggs and the blastula stage. The CgDy expression level was significantly(P<0.05)upregulated in the late trochophore(T2) and juvenile(J)stages compared with the gastrula,while umbo through spat had significantly lower expression than the gastrula(Fig. 5b).
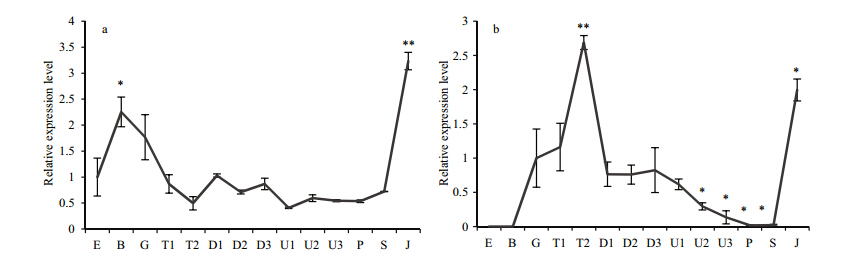 |
| Fig. 5 Expression pattern of CgDx(a) and CCgDy(b) transcripts in different developmental stages
RS18gene expression was used as an internal control. Egg and gastrula samples were used as the reference samples in the expression analysis of CgDx and CgDy,respectively. Vertical bars represent the SD. Differences determined as statistical significance are indicated by asterisks(* P<0.05,** P<0.01). E: egg; B: blastula; G: gastrula; T: trochophore; D: D-shape; U: umbo; P: pediveliger; S: spat; J: juvenile. |
CgDx transcripts were detected in all tested organs. The adductor muscle and mantle had significantly higher(P<0.05)expression levels than that of the digestive gl and ,gonad,hemolymph,gill, and labial palps(Fig. 6a). In contrast,the CgDytranscripts were undetectable in the mantle and adductor muscle. The highest expression level of CgDywas observed in the digestive gl and ,with 124.5-fold higher expression than that in the gill(which exhibited the lowest level of expression). CgDyexpression in the labial palps,gonad, and hemolymph was 97.2-,10.2-, and 5.3-fold higher than that of the gill(Fig. 6b).
 |
| Fig. 6 Expression pattern of CgDx(a) and CgDy(b) transcripts in different organs
EF gene expression was used as an internal control and a labial palps sample was used as the reference sample. Vertical bars represent the mean±SD(n=3). Lpa: labial palps; Gil: gill; Hae: hemolymph; Gon: gonad; Dgl: digestive gl and ; Man: mantle; Amu: adductor muscle. |
To investigate the regulation of oyster deiodinase synthesis,the expression profiles of CgDx and CgDyin T4 treatment and after EPI stimulation were determined by qRT-PCR,using RS18as a reference gene. In the T4 treatment,CgDx expression was significantly increased only at 24 h post treatment(hpt)(Fig. 7a). CgDyexpression increased at all-time points tested(Fig. 7b). After EPI stimulation,CgDx transcripts increased by 1.77-fold compared to the control at 3 h post stimulation(hps),then decreased by 0.65-fold at 6 hps, and increased again at 12 hps and 24 hps(Fig. 7c). The expression level of CgDyincreased only at 3 hps and did not differ significantly from the control at other time points(Fig. 7d).
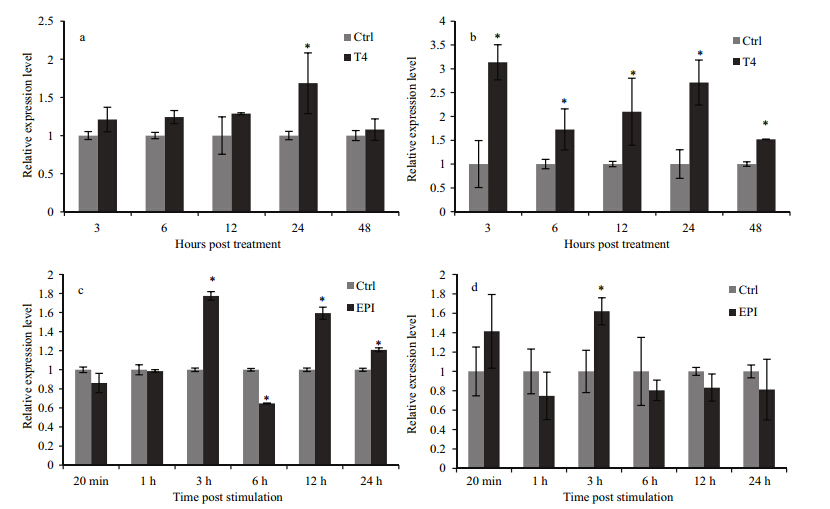 |
| Fig. 7 Temporal expression of CgDx(a,c) and CgDy(b,d) mRNA detected by qRT-PCR in oyster pediveliger larvae in T4 treatment(a,b)or after EPI stimulation(c,d)
RS18gene expression was used as an internal control. Data are normalized to the control group at each time point and displayed asthe mean±SD of duplicate independent experiments. A significant difference between the stimulation group and control group is indicated by asterisks(P<0.05). |
To investigate the origin and function of deiodinase genes,two iodothyronine deiodinases were cloned and characterized in C. gigas. The in-frame TGA codon and SECIS in the oyster deiodinase cDNA indicated that the oyster deiodinases were bona fide selenoproteins with a genuine catalytic activity center. A catalytically active deiodinase of the ascidian H. roretzihas been successfully expressed with its own SECIS in the HrDx 3′-UTR in mammalian cell lines(Shepherdley et al., 2003). Furthermore,amino acids essential for deiodinase activity were conserved in C. gigas(Fig. 3). The activity of CgDx and CgDy seems thus to be preserved. Actually,the scallop deiodinase(CfDx),the only deiodinase previously reported in non-chordate invertebrates,also had preserved activity of transforming T4 to T3,supported by an RNA interference experiment(Wu et al., 2012). Furthermore,we observed the possible transformation from T4 to T3 in oyster larvae in vivo(unpublished data). The two deiodinases were speculated to be responsible for this transformation from T4 to T3. However,it is unknown whether one or both are responsible, and further studies are needed to determine the zymologic characteristics.
Because only two invertebrate deiodinase sequences(HrDx and CfDx)were available,the origin and evolution of deiodinases in bilaterians were unclear. To determine the evolutionary relationships between vertebrate and invertebrate deiodinases,we searched the available invertebrate genomes for homologs. Because the in-frame TGA codons are predicted as stop codons by gene prediction software,selenoproteins have often escaped the attention of genome annotators(Lobanov et al., 2007). The selenoprotein-coding genes are usually inappropriately predicted as pseudogenes, and the deiodinases in the Pacific oyster genome have also been predicted as truncated pseudogenes. To avoid the gene prediction problem,a tblastn search was performed. As a result,the deiodinases were found in two other invertebrateorganisms,i. e.,the mollusk Lottia gigantea and the annelid Capitella teleta. Along with the two cloned oyster deiodinases,nine additional invertebrate deiodinases were identified in the study,which enabled the construction of a more reliable phylogenetic tree. The results revealed that the deiodinase gene originated from a common ancestor and a clade-specific gene duplication occurred independently during the differentiation of the mollusk,annelid, and vertebrate lineages. The duplication-derived vertebrate deiodinases were differentiated to D1,D2, and D3. Likewise,multiple duplication events have occurred independently during the evolution of mollusks and annelids(Fig. 4). As a result,the functions of those duplication-derived invertebrate deiodinases may not evolve a catalyzing specificity of ORD or IRD as described in vertebrate deiodinases. To investigate whether the function of the two oyster deiodinases diverged or was redundant,the spatiotemporal distribution of oyster deiodinase mRNAs were studied by qRT-PCR.
The spatiotemporal expression pattern could offer important clues to underst and gene function. The relatively stable expression of CgDx during ontogeny indicated that CgDx had a fundamental function throughout the development of the oyster. The CgDx mRNA transcripts were detected in the eggs,indicating that they could be maternally inherited in C. gigas. Maternally inherited D1 and THs of maternal origin were also reported in Xenopus laevis, and thus supported the speculation that TH signaling is involved in early amphibian development(MorvanDubois et al., 2008). We supposed that maternal THs were presented in eggs and TH signaling was also involved in early oyster development. The expression peaks of CgDx suggested that positive regulation of TH level by CgDx was increased in the blastula and juvenile stages. CgDymRNA levels showed a distinct and dynamic pattern across developmental stages,indicating that the function of CgDymay be more specific than that of CgDx . The expression patterns of CgDx and CgDyalso differed among organs. Correspondingly,the functionally differentiated vertebrate deiodinases(D1,D2, and D3)had distinct spatiotemporal expression patterns(Bianco et al., 2002). The spatiotemporal expression profiles of the two duplication-derived oyster deiodinases indicated functional divergence.
In vertebrates,TH homeostasis is maintained through a feedback regulation mechanism(Bianco et al., 2002),in which deiodinases either activate or inactivate THs and the transcription of deiodinases is in return regulated by TH status via TR. TR is a lig and -dependent transfactor that regulated gene expression through binding a specific DNA sequence(TRE)in the promoter of target genes(Zhang and Lazar, 2000). Lig and -coupled human TR demonstrates positive regulation of D1 and D3, and negative regulation of D2(Zavacki et al., 2005; Barca-Mayo et al., 2011)via TRE binding on the promoters of deiodinases(Jakobs et al., 1997; Nakajima et al., 2012). The expression of vertebrate deiodinases is regulated by THs and vertebrate deiodinases are TH responsive genes(Bianco et al., 2002). To investigate whether the oyster deiodinases were also regulate by THs,CgDx and CgDymRNA expression were measured during T4 treatment of pediveliger larvae. The results showed that the expression of both CgDx and CgDymRNA was induced. Moreover,atypical putative TREs(DR6 and DR1)were found in the promoter of the two deiodinases. Although,vertebrate TRs are known to specifically bind DR4,the reported invertebrate Schistosoma mansoniTRs can bind to DR0–DR5,which indicates that invertebrate TRs may not have evolved a distinct specificity with regard to spacing between half sites(Wu et al., 2007). It was possible that oyster TRs could bind to the atypical putative TREs(DR6 and DR1)of oyster deiodinases and then regulate CgDx and CgDyexpression. The expression of CgDx and CgDy was influenced by T4 treatment in oysters and TREs were found in their promoters,indicating that CgDx and CgDywere TH responsive genes. These results indicated that the oyster deiodinases were feedback regulated by TH and provided the first evidence for the existence of a TH feedback regulation mechanism in mollusks. The evolutionary origin of the TH system might date back to mollusks.
Thyroid hormone-regulated metamorphosis is a widespread feature in chordates, and is exemplified by the transformation of a tadpole into a frog(Tata,2006; Laudet,2011). THs also promote settlement and metamorphosis in two abalone species,H. discus and H. gigantean(Fukazawa et al., 2001). THs were thus may involve in metamorphosis of oysters. Meanwhile,EPI is a catecholamine hormone that effectively promotes oyster metamorphosis without settlement(Coon et al., 1985). To investigate whether the two hormones promoting metamorphosis were part of the same signaling pathway,the expression of two oyster deiodinases was monitored after EPI stimulation of metamorphosis-competent pediveliger larvae. The results showed that the expression of CgDx and CgDywere influenced by EPI stimulation,which indicated the possible involvement of CgDx and CgDyin the EPI initiated metamorphosis of oysters. The regulatory action of the adrenergic system on D2expression was also reported in rat brown adipose tissue,in which the D2expression level increases after EPI stimulation(Croteau et al., 1996). 5 CONCLUSION
In this study,two iodothyronine deiodinases were cloned in C. gigas. Our analysis revealed that the deiodinase gene originated from an common ancestor and a clade-specific gene duplication occurred independently during the differentiation of the mollusk,annelid, and vertebrate lineages. The expression of oyster deiodinases were feedback regulated by THs,suggesting the existence of a conserved TH feedback regulation mechanism in mollusks, and providing insights into TH evolution.
| Barca-Mayo O, Liao X H, Alonso M, Di Cosmo C, Hernandez A, Refetoff S, Weiss R E. 2011. Thyroid hormone receptor α and regulation of type 3 deiodinase. Mol. Endocrinol., 25 (4): 575-583. |
| Berry M J, Banu L, Harney J W, Larsen P R. 1993. Functional characterization of the eukaryotic secis elements which direct selenocysteine insertion at UGA codons. EMBO J., 12 (8): 3 315-3 322. |
| Bianco A C, Salvatore D, Gereben B, Berry M J, Larsen P R. 2002. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr. Rev., 23 (1): 38-89. |
| Coon S L, Bonar D B, Weiner R M. 1985. Induction of settlement and metamorphosis of the Pacific oyster, Crassostrea gigas (Thunberg), by l-Dopa and catecholamines. J. Exp. Mar. Biol. Ecol., 94 (1-3): 211- 221. |
| Croteau W, Davey J C, Galton V A, St Germain D L. 1996. Cloning of the mammalian type II iodothyronine deiodinase. A selenoprotein differentially expressed and regulated in human and rat brain and other tissues. J. C lini. I nvest., 98 (2): 405-417. |
| Das B, Heimeier R A, Buchholz D R, Shi Y B. 2009. Identification of direct thyroid hormone response genes reveals the earliest gene regulation programs during frog metamorphosis. J. Biol. Chem., 284 (49): 34 167-34 178. |
| Du Y S, Zhang L L, Xu F, Huang B Y, Zhang G F, Li L. 2013. Validation of housekeeping genes as internal controls for studying gene expression during Pacific oyster (Crassostrea gigas) development by quantitative realtime PCR. Fish Shellfish Immunol., 34 (3): 939-945. |
| Fukazawa H, Hirai H, Hori H, Roberts R D, Nukaya H, Ishida H, Tsuji K. 2001. Induction of abalone larval metamorphosis by thyroid hormones. Fisheries Sci., 67 (5): 985-987. |
| Heyland A, Price D A, Bodnarova-Buganova M, Moroz L L. 2006. Thyroid hormone metabolism and peroxidase function in two non-chordate animals. J. Exp. Zool. B Mol. Dev. Evol., 306B (6): 551-566. |
| Jakobs T C, Schmutzler C, Meissner J, Köhrle J. 1997. The promoter of the human type I 5'-deiodinase gene— mapping of the transcription start site and identification of a DR+4 thyroid-hormone-responsive element. Eur. J. Biochem., 247 (1): 288-297. |
| Köhrle J. 2002. Iodothyronine deiodinases. Methods Enzymol., 347 : 125-167. |
| Laudet V. 2011. The origins and evolution of vertebrate metamorphosis. Curr. Biol., 21 (18): R726-R737. |
| Lobanov A V, Fomenko D E, Zhang Y, Sengupta A, Hatfield D L, Gladyshev V N. 2007. Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol., 8 (9): R198. |
| Mariotti M, Lobanov A V, Guigo R, Gladyshev V N. 2013. SECISearch3 and Seblastian: new tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res., 41 (15): e149. |
| Morvan-Dubois G, Demeneix B A, Sachs L M. 2008. Xenopus laevis as a model for studying thyroid hormone signalling: From development to metamorphosis. Mol. Cell. Endocrinol., 293 (1-2): 71-79. |
| Nakajima K, Fujimoto K, Yaoita Y. 2012. Regulation of thyroid hormone sensitivity by differential expression of the thyroid hormone receptor during Xenopus metamorphosis. Genes Cells, 17 (8): 645-659. |
| Qu T, Huang B Y, Zhang L L, Li L, Xu F, Huang W, Li C Y, Du Y S, Zhang G F. 2014. Identification and functional characterization of two executioner caspases in Crassostrea gigas. PloS One, 9 (2): e89040. |
| Roustaian P, Gaik L A. 2006. Effect of thyroxine immersion on larval survival, growth and postlarvae production of freshwater prawn, Macrobrachium rosenbergii (de Man). Aquac. Res., 37 (13): 1 378-1 380. |
| Schweizer U, Schlicker C, Braun D, Kohrle J, Steegborn C. 2014. Crystal structure of mammalian selenocysteinedependent iodothyronine deiodinase suggests a peroxiredoxin-like catalytic mechanism. Proc. Natl. Acad. Sci. USA, 111 (29): 10 526-10 531. |
| Shepherdley C A, Klootwijk W, Makabe K W, Visser T J, Kuiper G G J M. 2003. An ascidian homolog of vertebrate iodothyronine deiodinases. Endocrinology, 145 (3): 1 255-1 268. |
| Tata J R. 2006. Amphibian metamorphosis as a model for the developmental actions of thyroid hormone. Mol. Cell. Endocrinol., 246 (1-2): 10-20. |
| Wu T T, Shi X W, Zhou Z, Wang L L, Wang M Q, Wang L L, Huang M M, Yang C Y, Song L S. 2012. An iodothyronine deiodinase from Chlamys farreri and its induced mRNA expression after LPS stimulation. Fish Shellfish Immunol., 33 (2): 286-293. |
| Wu W J, Niles E G, Loverde P T. 2007. Thyroid hormone receptor orthologues from invertebrate species with emphasis on Schistosoma mansoni. BMC Evol. Biol., 7 : 150. |
| Yen P M. 2001. Physiological and molecular basis of thyroid hormone action. .Physiol. Rev, 81 (3): 1 097-1 142. |
| Zavacki A M, Ying H, Christoffolete M A, Aerts G, So E, Harney J W, Cheng S Y, Larsen P R, Bianco A C. 2005. Type 1 iodothyronine deiodinase is a sensitive marker of peripheral thyroid status in the mouse. Endocrinology, 146 (3): 1 568-1 575. |
| Zhang G F, Fang X D, Guo X M, Li L, Luo R B, Xu F, Yang P C, Zhang L L, Wang X T, Qi H G, Xiong Z Q, Que H Y, Xie Y L, Holland P W H, Paps J, Zhu Y B, Wu F C, Chen Y X, Wang J F, Peng C F, Meng J, Yang L, Liu J, Wen B, Zhang N, Huang Z Y, Zhu Q H, Feng Y, Mount A, Hedgecock D, Xu Z, Liu Y J, Domazet-Lošo T, Du Y S, Sun X Q, Zhang S D, Liu B H, Cheng P Z, Jiang X T, Li J, Fan D D, Wang W, Fu W J, Wang T, Wang B, Zhang J B, Peng Z Y, Li Y X, Li N, Wang J P, Chen M S, He Y, Tan F J, Song X R, Zheng Q M, Huang R L, Yang H L, Du X D, Chen L, Yang M, Gaffney P M, Wang S, Luo L H, She Z C, Ming Y, Huang W, Zhang S, Huang B Y, Zhang Y, Qu T, Ni P X, Miao G Y, Wang J Y, Wang Q, Steinberg C E W, Wang H Y, Li N, Qian L M, Zhang G J, Li Y R, Yang H M, Liu X, Wang J, Yin Y, Wang J. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature, 490 (7418): 49-54. |
| Zhang J S, Lazar M A. 2000. The mechanism of action of thyroid hormones. Annu. Rev. Physiol., 62 : 439-466. |
| Zhang L L, Li L, Zhang G F. 2011. Gene discovery, comparative analysis and expression profile reveal the complexity of the Crassostrea gigas apoptosis system. Dev. Comp. Immunol., 35 (5): 603-610. |
 2015, Vol. 33
2015, Vol. 33



