Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CAO Cuicui(曹翠翠), LIU Miao(刘淼), GUO Shaoru(郭少茹), ZHAO Dan(赵丹), LUAN Rixiao(栾日孝), WANG Hongwei(王宏伟)
- Grateloupia ramosa Wang & Luan sp. nov.(Halymeniaceae, Rhodophyta), a new species from China based on morphological evidence and comparative rbcL sequences
- Chinese Journal of Oceanology and Limnology, 34(2): 283-294
- http://dx.doi.org/10.1007/s00343-015-4335-z
Article History
- Received Nov. 24, 2014
- accepted in principle Mar. 17, 2016
2 Dalian Natural History Museum, Dalian 116023, China
The genus Grateloupia was established based on generitype G . filicina (Lamouroux) C. Agardh from the Mediterranean Sea (Agardh, 1822). This genus is highly diverse, consisting of more than 90 species distributed worldwide in cold-temperate to tropical waters (Wang et al., 2000, 2001; Kawaguchi et al., 2001; Gavio and Fredericq, 2002; Faye et al., 2004; De Clerck et al., 2005a, b; Wilkes et al., 2005; Lin et al., 2008; Zhao et al., 2012; Guiry and Guiry, 2013). To date, 36 Grateloupia species have been reported in China (Xia, 2004; Zhang et al., 2012; Zhao et al., 2012; Yu et al., 2013; Liu et al., 2014; Wang et al., 2015). Among these, G . orientalis S.-M. Lin & H.-Y. Liang is a new record from Hainan, China (Zhang et al., 2012), and G . dalianensis, G . yinggehaiensis, G . tenuis, G . boaoensis and G . huanghaiensis are recently reported species (Zhang et al., 2012; Yu et al., 2013; Liu et al., 2014; Wang et al., 2015). Classification of members of Grateloupia is extremely difficult because differences between the external morphology and internal structure have prevented development of uniform morphological classification criteria. Gargiulo et al. (2013) recently considered the formation of carpogonial and auxiliary cell ampullae and post fertilization events to be important characteristics for future generic reassessment of Grateloupia . Additionally, they found that the genus was strongly supported by rbc L phylogenies. Overall, they divided Grateloupia into eight subgroups, G . lanceolata -group, G . americana -group, G . belangeri -group, G . phuquocensis -group, G . stipitata -group, G . filicina -group, G . doryphora - group and G . montagnei -group.
G . catenata was first described in 1920 (Yendo, 1920), after which Howe (1924) described G . filicina (Lamouroux) C. Agardh var. lomentaria Howe with no reference to Yendo's (1920) publication. Okamura (1936) considered G . fllicina var. lomentaria to be a mature stage of G . filicina var. porracea and proposed combination with G . filicina var. porracea f. lomentaria (Howe) Okamura. At the same time, Okamura (1936) reduced G . catenata to synonymy with G . filicina var porracea f. lomentaria . Wang et al., (2000) then reinstated G . catenata based on morphological observations and rbc L analysis. G . tenuis was similar to G . catenata based on external morphology; however, Yu et al. (2013) confirmed a new species of G . tenuis through morphological observations and molecular phylogenetic studies.
The specimens investigated in the present study, which had an external morphology similar to G . catenata and G . tenuis, were collected from Lingshui (110°1′E, 18°30′N) and Wenchang (110°47′E, 19°32′N) of Hainan Province, southern China. The external and internal morphologies were distinguished from other species (G . tenuis, G . catenata, G . filicina, G . ramosissima, G . orientalis) for the first time based on observations. We also conducted rbc L sequence analysis to support the results of morphological observations. Herein, we propose that this alga be classified in the Grateloupia genus with the name Grateloupia ramosa Wang & Luan sp. nov.
2 MATERIAL AND METHOD 2.1 Morphological analysisSpecimens used in the present study were handcollected from Wenchang, Hainan Province, China. Voucher specimens have been deposited in the the Herbarium of the College of Life Sciences, Liaoning Normal University, Dalian, China (LNU) (1) (9 January 2009, leg . R. X. Luan; LNU20092016- LNU20092022), and (2) Lingshui, Hainan Province, China (5 February 2009, leg . R. X. Luan; LNU20092083-LNU20092088). We regarded G . ramosa (LNU20092086) as the holotype.
Morphological observations were made based on specimens preserved in 10% formalin/seawater using an anatomical lens or microscopic observation of several algae that contained the reproductive organs of algal specimens (LNU20092019-LNU200902020; LNU20092087-LNU20092089). To accomplish this, samples were selected, frozen, cut into small pieces of uniform size of about 1 cm, and placed in a petri dish. Photographs of the holotype specimen were taken with a Canon EOS 650D (Canon, Japan). Additionally, sections were made by cryostat microtome, stained with 0.5% (w/v) cotton blue and photographed using an Olympus BH 2 digital camera (Olympus Beijing Co. Ltd., China) mounted on a Nikon microscope (Nikon Corporation, Japan). The branch features, lateral branch formation, and cystocarp distribution were scanned using an anatomical lens.
2.2 Molecular analysisDNA of LNU20092016, LNU20092017, LNU20092018, LNU20092083, LNU20092084, and LNU20092085 was isolated using a DNeasy Plant Mini Kit (QIAGEN, Valencia, CA, USA). Sequences of 41 additional related red algal species were selected from GenBank and included in the alignments (Table 1). DNA extraction and sequencing were performed as previously described (Wang et al., 2000). PCR (Applied Biosystems, USA) was conducted in a 50 μL reaction mixture that consisted of 12.5 μL of 2×Taq PCR Master Mix, 31.5 μL of deionized water, 4 μL of template DNA, and 1 μL of each primer. PCR was carried out by subjecting the samples to 93℃ for 1 min, followed by 35 cycles of amplification (denaturation at 94℃ for 30 s, annealing at 53℃ for 30 s and extension at 72℃ for 45 s) and then final extension at 72℃ for 5 min.
|
The rbc L sequences for phylogenetic analyses were aligned using Clustal X version 1.83 (Thompson et al., 1997). Phylogenetic trees were constructed using the Bayesian analysis (BA) Maximum Likelihood (ML), Maximum Parsimony (MP) and Neighbor-Joining (NJ) methods with PAUP version 4.0 (Swofford, 2003).
For MP, all sites were considered unordered and equally weighted. The heuristic search option was used, with 100 random sequence additions and treebisection and reconnection branch exchanging. For this analysis, missing data were treated as gaps. The ML and NJ analyses were estimated using the hierarchical likelihood ratio test. To obtain the optimal settings and ensure data reliability, we used Modeltest version 3.7 (Posada and Buckley, 2004) evolutionary analyses in MEGA5 (Tamura et al., 2011). All complex models of molecular evolution were evaluated using the methods outlined by Litaker et al. (1999) and Moncalvo et al. (2000). Node supports were estimated using the bootstrap proportion values for calculation (Felsenstein, 1985) using ML (1 000 bootstrap resamplings), NJ (1 000 bootstrap resamplings) and MP (1 000 bootstrap resamplings) methods.
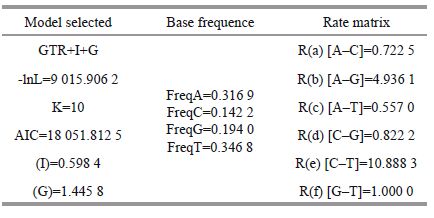
For Bayesian inference (BI), the dataset was partitioned according to codon positions and the prior probability distribution for the site-specific rates in the phylogenetic model was set as “variable”. Models of nucleotide substitution were estimated from Modeltest 3.7 (Posada and Buckley, 2004). Bayesian inference (BI) was performed with MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003) using the GTR+I+G (Table 2). The BI results yielded 10 000 trees, 25% of which were burni The remaining trees were used to calculate the consensus tree.
|
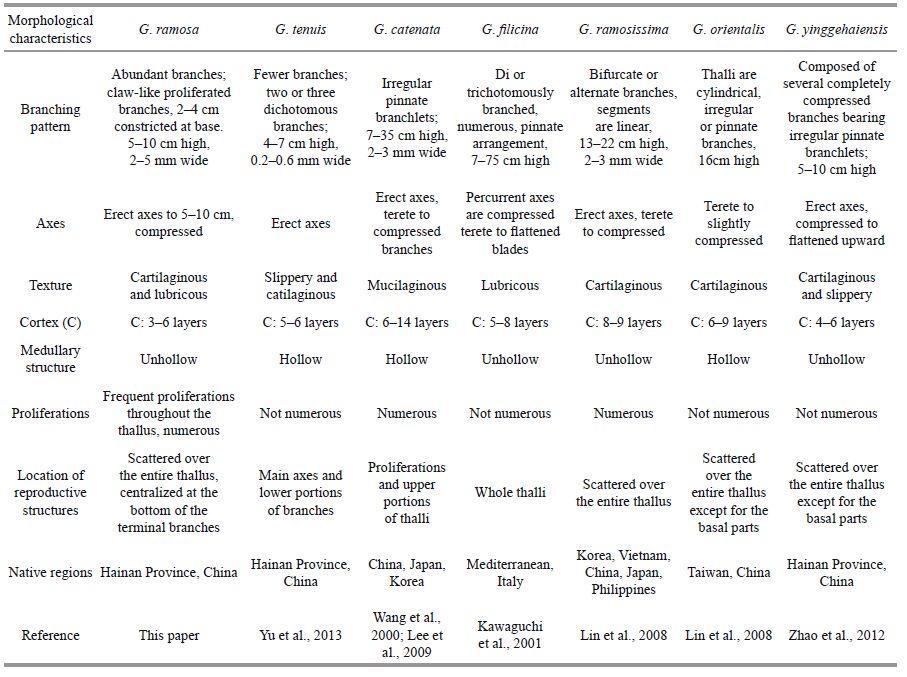
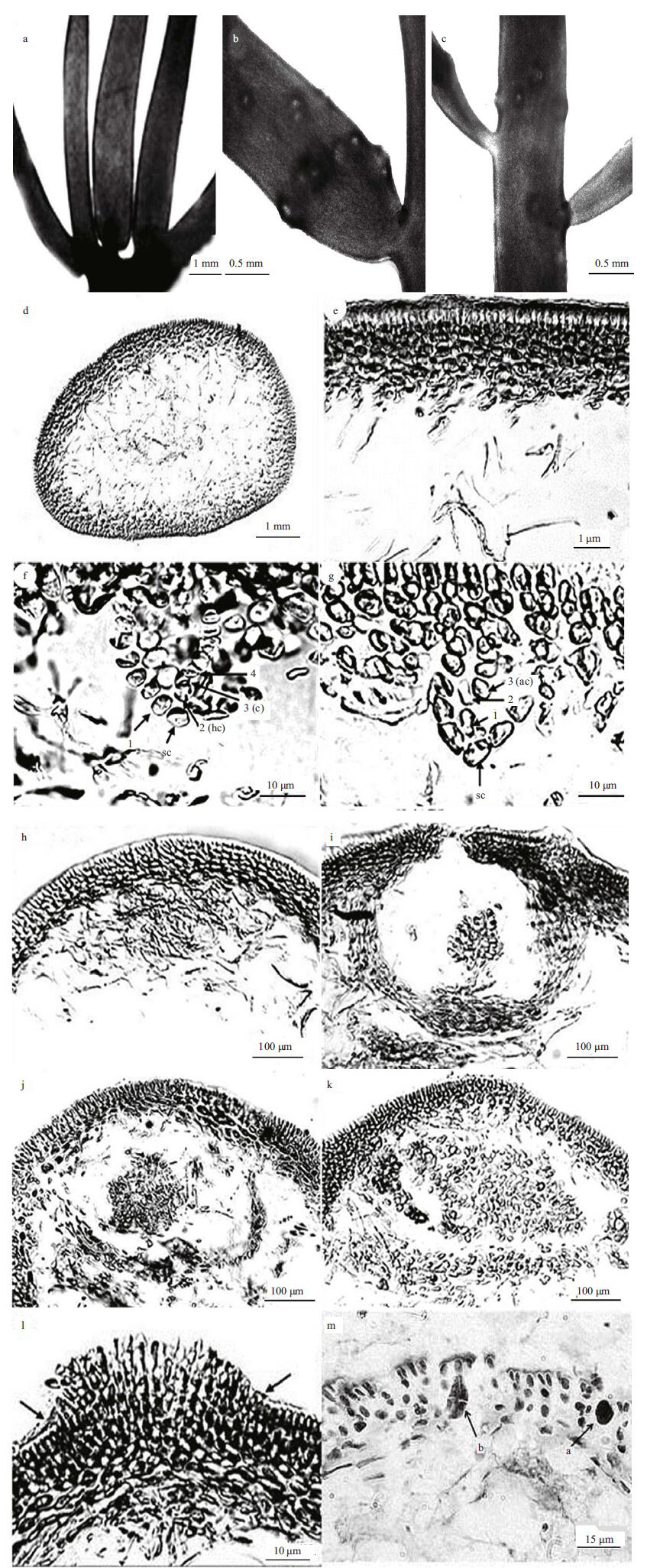
Thalli were 5-10 cm in height, purplish red in color, cartilaginous and lubricous in texture. Blades had numerous branches and an opposite arrangement . Percurrent axes were compressed (Fig. 1). Terminal branches consisted of up to five multiple claw-like branches that gradually tapered to the upper part (Fig. 2a). The lateral branchlets were constricted to 2-4 mm at the base, frequently with proliferations from the margins and the surfaces (Fig. 2b). The cortex consisted of 3-6 layers of elliptic or anomalous cells and a medulla composed of dense filaments. Gametophytes dioecious, reproductive structures distributed throughout the thallus and centralized at the bottom of the terminal branches (Fig. 2c). Spermatangia were developed from cortical cells of male gametophytes. Mature spermatangia were ellipsoidal, measuring 4- 6 mm in length and 3-4 mm in diameter. Carposporangia were produced from most cells of gonimoblasts and deeply immersed in the medulla. Mature cystocarps were spherical, 230- 350 μm in diameter. Tetrasporangia were divided cruciately, 35-45 μm long and 20-25 μm in diameter, and embedded in the outer cortex.

|
| Figure 1 Holotype specimen of G . ramosa (female gametophyte collected from Lingshui, LNU20092086) |
|
| Figure 2 Branching characteristics of G . ramosa, distribution of cystocarps, and transverse sections and reproductive structure of G . ramosa |
ETYMOLOGY: “ ramosa ” in reference to the numerous branches of thalli.
HOLOTYPE: Fig. 1 A female specimen (LNU20092086).
TYPE LOCALITY: Lingshui, Hainan Province (108.7°E, 18.5°N), southern China, 5 February 2009 by R. X. Luan.
DISTRIBUTION: At present, known only in Hainan Province, Southern China (108.7°E, 18.5°N).
Transverse sections of the thallus displayed a cortex of 3-6 layers of elliptical or anomalous cells, and a medulla consisting of compact medullary filaments 2-7 μm in diameter that ran in various directions. The medulla cells approached the inner cortical layer, which consisted of rounded or irregularly shaped cells (Fig. 2d). Spermatangia developed from the outer cortex cells of the male gametophytes (Fig. 2e). Mature spermatangia were ellipsoidal, measuring 4-6 μm in length and 3-4 μm in diameter. Both the carpogonial ampullae and auxiliary cell ampullae were initiated in the inner cortex. Specimens had a four-celled carpogonial branch borne on a supporting cell consisting of a hypogynous cell and a carpogonium (Fig. 2f). The hypogynous cell (hc) bore a three-celled lateral branch. An oval cell (4) bore a five-celled lateral branch, with another cell (1) bearing a six-celled lateral branch. The four-celled carpogonial branch and the lateral branches constituted an ampulla. Auxiliary-cell ampullae were Grateloupia -type. Auxiliary cells were cup-shaped and remarkably ellipsoidal or ovate in shape. The auxiliary cell was surrounded by the cup-shaped auxiliary-cell branch and contained a three-celled branch. The gonimoblast initium was generally cut off from the fertilized body and gave rise to gonimoblast filaments. Carposporangia, which were the most developed gonimoblasts cells, were deeply immersed in the medulla. Cystocarps were inconspicuous and surrounded by dense clusters of cells. Cystocarp development is shown (Fig. 2h-j). Mature cystocarps were spherical, with a diameter ranging from 400- 600 μm (Fig. 2j). Tetrasporangia were cruciately divided, 35-45 μm long, 5-15 μm in diameter, and embedded in the outer cortex (Fig. 2k).
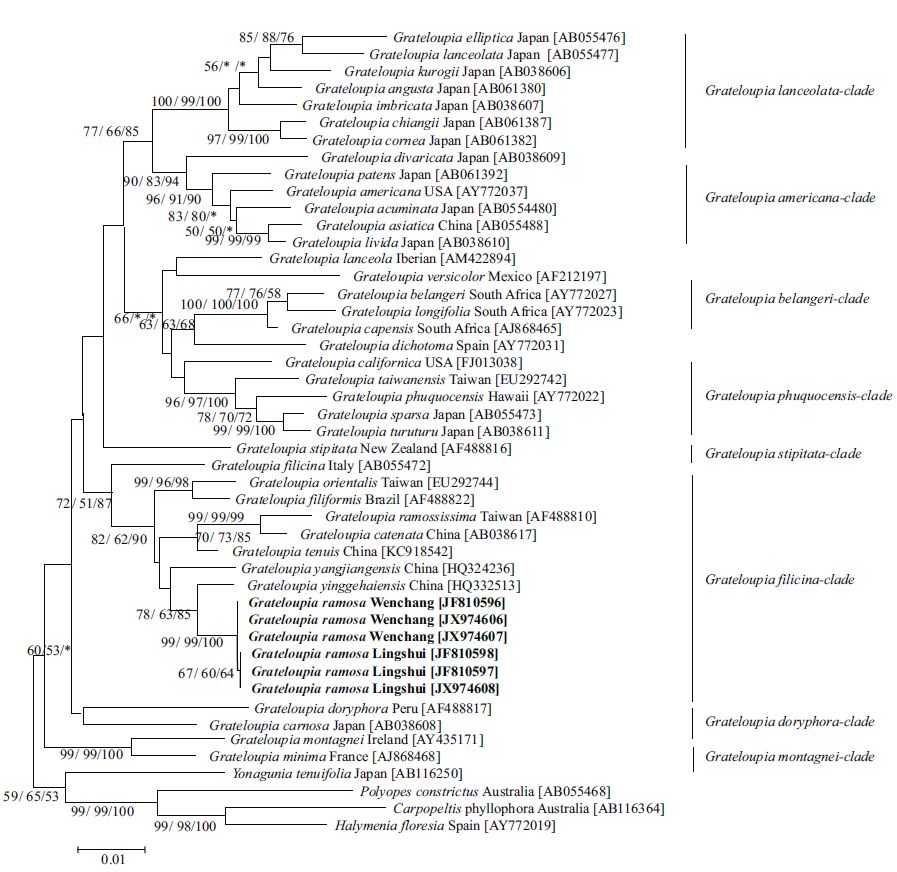
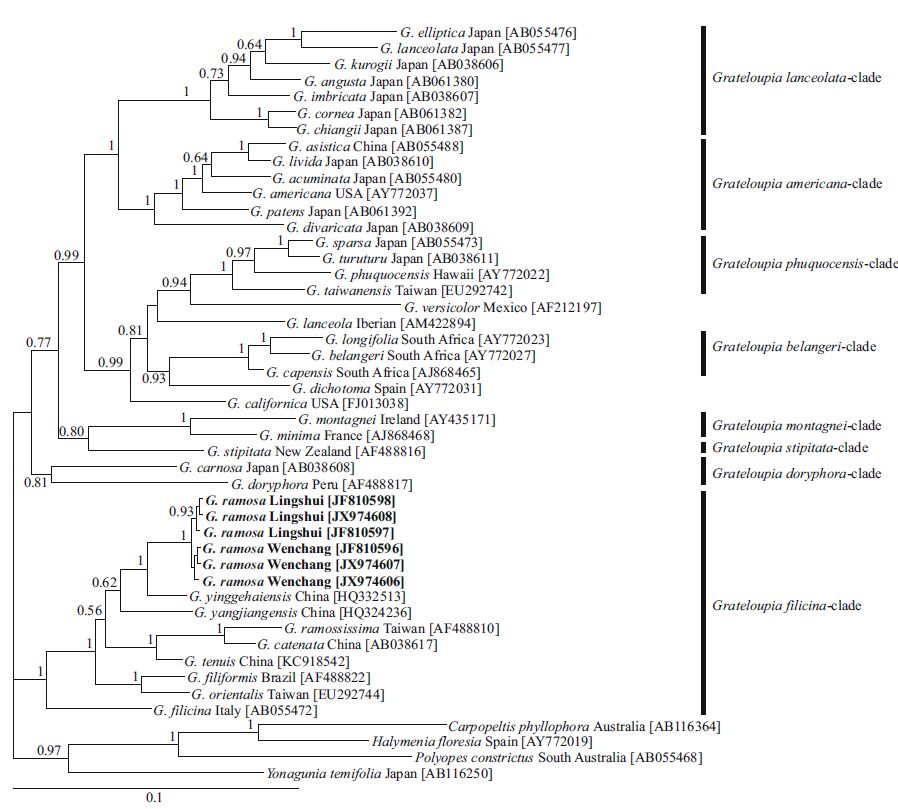
3.2 rbc L analysesWe compared the rbc L sequences of the six G . ramosa samples to the 37 selected species within Grateloupia and four species in other genera within Halymeniaceae (Figs.3, 4). Partial sequences of 1 251 base pairs (bp) corresponding to positions 101 to 1 351 of the full rbc L gene (1 467 bp) were used for the alignment.
Through rbc L gene sequence comparison, sequences from three samples, including the holotype of G . ramosa from the type locality of Lingshui, were identical, while the other three samples from Wenchang differed by only one nucleotide substitution. The pairwise divergence of these samples ranged from 0.0% to 0.08% (0-1 bp); thus, they can be treated as the same species. There were 62 or 63 bp changes (5.17%-5.26%) between G . ramosa and the generitype G . filicina . Additionally, there were 27- 28 bp changes (2.19%-2.28%) between G . ramosa and G . yinggehaiensis H. W. Wang & R. X. Luan, which was the closest relative to G . ramosa in all three phylogenetic trees. Finally, 44-45 bp changes (3.62%-3.71%) were observed between G . ramosa and G . catenata, which was most similar to G . ramosa in appearance. The pairwise distances between G . ramosa and G . ramosissima Okamura ranged from 56-57 bp changes (4.65%-4.74%), while there were 39-40 bp changes (3.12%-3.20%) between G . ramosa and G . tenuis Wang & Luan and between G . ramosa and G . orientalis S.-M. Lin & H.-Y. There were 48- 49 bp changes (3.96%-4.04%) in Liang, and these two species were within the same subclade as our species. When compared with the outgroup species of Yonagunia tenuifolia Kawaguchi & Masuda, Carpopeltis phyllophora (J. D. Hooker and Harvey) F. Schmitz, Halymenia floresia (Clemente) C. Agardh and Polyopes constrictus (Turner) J. Agardh, the pairwise distances ranged from 106-132 bp changes (9.01%-11.49%).
The NJ, MP, ML and Bayesian phylogenetic trees are presented in Figs.3, 4, respectively. There were two main clades in all phylogenetic trees, Grateloupia clade and outgroups within the Halymeniaceae clade. In addition, there are eight subclades and several undefined branches of the selected species within the large Grateloupia sensu lato clade. The G . ramosa specimens formed an individual subclade in the G . filicina -clade within the large Grateloupia sensu lato clade. Finally, sister species to our species in the G . filicina -clade consisted of G . yangjiangensis, G . orientalis, G . filiformis, G . yinggehaiensis, and G . tenuis, with G . yinggehaiensis being most closely related to our species.
4 DISCUSSIONCharacteristics of the auxiliary-cell ampullae and external morphology are used for the generic classification of Halymeniaceae. Chiang (1970) found that the developmental processes of auxiliary-cell ampullae and carpogonial branch ampullae were relatively stable, and were an important boundary for different genera within the Halymeniaceae. Gargiulo et al. (2013) reported almost all of the types of carpogonial and auxiliary cell ampullae within Halymeniaceae, while Lin et al. (2008) reported two different types of G . taiwanensis and G . orientalis . The auxiliary-cell ampullae types in Halymeniaceae include Aeodes, Cryptonemia, Halymenia, Grateloupia and Thamnoclonium . The Grateloupia - type structure is simplest, containing a single primary ampullary filament that produces two or three 7-13-celled unbranched secondary amlullary filaments. Chiang (1970) reported that six genera in Halymenniaceae (Grateloupia C. Agardh, Prionitis J. Agardh, Pachymeniopsis Yamada ex Kawabata, Dermocorynus P. L. Crouan & H. M. Crouan, Yonagunia Kawaguchi & Masuda and Phyllymenia J. Agardh) had Grateloupia -type amlullae (Chiang, 1970; Kawaguchi et al., 2004; Mateo-Cid et al., 2005; Gargiulo et al., 2013). Wang et al. (2001) transferred Pachymeniopsis and Prionitis to Grateloupia (Kawaguchi, 1997; Wang et al., 2001), and Phyllymenia has also been merged to Grateloupia (De Clerck et al., 2005b). The genus Sinotubimorpha also had simple Grateloupia -type ampullae, and was therefore transferred to Grateloupia after ampullary morphological observation and molecular analysis (Sheng et al., 2012). However, Gargiulo et al. (2013) recently resurrected the four groups based on the architecture of carpogonial and auxiliary cell ampullae and the early postfertilization events. Corresponding to Chiang (1970), Gargiulo et al. (2013) and other authoritative algal taxonomists investigating Grateloupia species (Wang et al., 2000; Faye et al., 2004; Lin et al., 2008; Gargiulo et al., 2013), the architecture of carpogonial and auxiliary cell ampullae of our species (Fig. 2f, g) shows that it is a member of the family Halymeniaceae that belongs to the genus Grateloupia based on its four-celled carpogonial branch and 3-celled auxiliary branch.
|
| Figure 3 Maximum likehood (ML) tree based on partial rbc L gene sequences data showing the phylogenetic relationship of Grateloupia ramosa with the other species within Grateloupia and species in other related genera in Halymeniaceae |
|
| Figure 4 Bayesian tree based on partial rbc L gene sequences data showing the phylogenetic relationship of Grateloupia ramosa with the other species within Grateloupia and species in other related genera in Halymeniaceae |
The proposed G . ramosa species is characterized by its linear thalli, median height of 5-10 cm, percurrent compressed axes, abundant branches that are opposite in arrangement, and constriction at the base. The terminal branches consisted of multiple claw-like branches gradually tapering to the apices. Reproductive structures are distributed throughout the thallus, especially centralize the bottom of the terminal branches. Table 3 shows a comparison of morphological characteristics of G . ramos a and the nearby species, G . catenata, G . filicina, G . ramosissima, G . orientalis and G . tenuis . The results demonstrate that the features of this new species differed from other species, including the generitype G . filicina . G . yinggehaiensis is the most closely related to our species and has the same type of ampullae. However, these organisms are not similar morphologically. Specifically, G . yinggehaiensis is foliated and composed of several completely compressed branches bearing irregular pinnate branchlets, whereas G . ramosa is linear, with percurrent axes bearing vimineous claw-like branchlets. Prominent morphological diversity was also observed between G . ramosa and G . catenata . These species are somewhat similar based on their outer appearance and numerous proliferations, but have differing branching patterns, habits, and locations of reproductive structures and the medulla. The erect axes of G . catenata are up to 35 cm high. The erect axes are terete below, becoming compressed up to 2 mm wide above (Wang et al., 2000), whereas our species has shorter compressed erect axes 5-10 cm high. The proliferations arising from G . catenata 's erect axes have one or two constrictions (Wang et al., 2000), but our species only has claw-like proliferations and constrict at the base. G . catenata has a hollow medulla, but the medulla of G . ramosa has compact medullary filaments. These data indicated that the studied species was not identical to G . catenata .
The molecular phylogenetic data also support the declaration of a new Grateloupia species. These data show 27-28 bp (2.19%-2.28%) divergence between G . ramosa and G . yinggehaiensis, which was closest to G . ramosa in the phylogenetic tree, even though these species are not at all similar morphologically (Table 3). Moreover, the pairwise distance between species of Grateloupia was 13-111 bp (1.0%-8.8%) (Kawaguchi et al., 2002), indicating that the proposed species belonged within the Grateloupia and was not the same species as G . yinggehaiensis . There was also 44-45 bp (3.62%-3.71%) divergence between G . ramosa and G . catenata, which was most similar to the proposed species in external morphology. G. ramosa and G . tenuis showed a 39-40 bp difference, but were similar in external morphology. These sequence divergences were well within the interspecific values observed within Grateloupia (García-Jiménez et al., 2008; Lee et al., 2009). The sequence divergence between our species and outgroups in other genera within Halymeniaceae [106-132 bp (9.01%-11.49%)] clearly indicate that G . ramosa belongs to the genus Grateloupia, family Halymeniaceae. The phylogenetic tree also clearly revealed eight subclades of the selected species within the large Grateloupia sensu lato clade, G . lanceolata - clade, G . americana -clade, G . belangeri -clade, G . phuquocensis -clade, G . stipitata -clade, G . filicina - clade, G . doryphora -clade and G . montagnei -clade. The G . ramosa specimens formed an individual subclade in the G . filicina -clade. The three samples collected from Wenchang and three from Lingshui, which are both in Hainan Province, China, only had 1 bp divergences and could therefore be considered one subclade within the G . filicina -clade with the sister related species G . yangjiangensis, G . orientalis, G . filiformis, G . tenuis and G . yinggehaiensis, among which G . yinggehaiensis was most closely related to our species.
The conclusions obtained in the paper support the point of view suggested by Gargiulo et al. (2013); thus, based on morphological and rbc L sequence data, it is proposed that these specimens belong to the genus Grateloupia and should be named G . ramosa Wang & Luan sp. nov.
5 ACKNOWLEDGEMENTWe thank SHENG Yingwen, ZHANG Wen, and SHI Chunguang for providing technical assistance.
| Agardh C A, 1822. Species algarum rite cognitae. vol. 1. part 2. Lundae, Lund, Sweden.[I-VⅢ] p : 169 –389. |
| Chiang Y M, 1970. Morphological studies of red algae of the family Cryptonemiaceae. University of California Publications in Bot any, 58 : 1 –95. |
| De Clerck O, Gavio B, Fredericq S, Bárbara I, Coppejans E, 2005a. Systematics of Grateloupia filicina(Halymeniaceae, Rhodophyta), based on rbcL sequence analyses and morphological evidence, including the reinstatement of G. minima and the description of G.capensis sp. nov. Journal of Phycology, 41 (2) : 391 –410. Doi: 10.1111/(ISSN)1529-8817 |
| De Clerck O, Gavio B, Fredericq S, Cocquyt E, Coppejans E, 2005b. Systematic reassessment of the red algal genus Phyllymenia(Halymeniaceae, Rhodophyta). Eur opean J ournal of Phycology, 40 (2) : 169 –178. Doi: 10.1080/09670260500128343 |
| Faye E J, Wang H W, Kawaguchi S, Shimada S, Masuda M, 2004. Reinstatement of Grateloupia subpectinata(Rhodophyta, Halymeniaceae) based on morphology and rbcL sequences. Phycological Res earch, 52 (1) : 59 –67. Doi: 10.1111/pre.2004.52.issue-1 |
| Felsenstein J, 1985. Confidence limits on phylogenies:an approach using the bootstrap. Evolution, 39 (4) : 783 –791. Doi: 10.2307/2408678 |
| García-Jiménez P, Geraldino P J L, Boo S M, Robaina R R, 2008. Red alga Grateloupia imbricata(Halymeniaceae), a species introduced into the Canary Islands. Phycol ogical Res earch, 56 (3) : 166 –171. Doi: 10.1111/pre.2008.56.issue-3 |
| Gargiulo G M, Morabito M, Manghisi A, 2013. A re-assessment of reproductive anatomy and postfertilization development in the systematics of Grateloupia(Halymeniales, Rhodophyta). Cryptogamie, Algologie, 34 (1) : 3 –35. Doi: 10.7872/crya.v34.iss1.2013.3 |
| Gavio B, Fredericq S, 2002. Grateloupia turuturu(Halymeniaceae, Rhodophyta) is the correct name of the non-native species in the Atlantic known as Grateloupia doryphora. Eur opean J ournal of Phycol ogy, 37 (3) : 349 –359. |
| Guiry M D, Guiry G M. 2013. AlgaeBase, www electronic publication. National University of Ireland, Galway. http://www.algaebase.org. Searched on 8 January 2013. |
| Howe M A, 1924. Chinese marine algae. Bulletin of Torrey Botanical Club, 51 (4) : 133 –144. Doi: 10.2307/2480234 |
| Kawaguchi S, Shimada S, Wang H W, Masuda M, 2004. The new genus Yonagunia Kawaguchi et Masuda(Halymeniaceae, Rhodophyta), based on Y. tenuifolia Kawaguchi et Masuda sp. nov. from southern Japan and including Y. formosana(Okamura) Kawaguchi et Masuda comb. nov. from southeast Asia. Journal of Phycology, 40 (1) : 180 –192. |
| Kawaguchi S, Wang H W, Horiguchi T, Lewis J A, Masuda M, 2002. Rejection of Sinkoraena and transfer of Some species of Carpopeltis and Sinkoraena y to Polyopes(Rhodophyta, Halymeniaceae). Phycologia, 41 (6) : 619 –635. Doi: 10.2216/i0031-8884-41-6-619.1 |
| Kawaguchi S, Wang H W, Horiguchi T, Sartoni G, Masuda M, 2001. A comparative study of the red alga Grate loupia filicina(Halymeniaceae) from the northwestern Pacific and Mediterranean with the description of Grate loupia asiatica sp nov.. Journal of Phycology, 37 (3) : 433 –442. Doi: 10.1046/j.1529-8817.2001.037003433.x |
| Kawaguchi S, 1997. Taxonomic notes on the Halymeniaceae(Gigartinales, Rhodophyta) from Japan. Ⅲ.Synonymization of Pachymeniopsis Yamada in Kawabata with Grateloupia C. Agardh. Phycol ogical Res earch, 45 (1) : 9 –21. Doi: 10.1111/pre.1997.45.issue-1 |
| Lee J I, Kim H G, Geraldino P J L, Hwang I K, Boo S M, 2009. Molecular classification of the genus Grateloupia(Halymeniaceae, Rhodophyta) in Korea. Algae, 24 (4) : 231 –238. Doi: 10.4490/ALGAE.2009.24.4.231 |
| Lin S M, Liang H Y, Max H H, 2008. Two types of auxiliary cell ampullae in Grateloupia(Halymeniaceae, Rhodophyta), including G. taiwanensis sp. nov. and G.orientalis sp. nov. from Taiwan based on rbcL gene sequence analysis and cystocarp development. Journal of Phycology, 44 (1) : 196 –214. Doi: 10.1111/jpy.2008.44.issue-1 |
| Litaker R W, Tester P A, Colorni A, Levy M G, Noga E J, 1999. The phylogenetic relationship of Pfiesteria piscicida, cryptoperidiniopsoid sp. Amyloodin o um ocellatum and a Pfiestera-like dinoflagellate to other dinoflagellates and apicomplexans. Journal of Phycolog y, 35 (6) : 1379 –1389. Doi: 10.1046/j.1529-8817.1999.3561379.x |
| Liu M, Wang H W, Luan R X, 2014. Morphological observation and rbcL sequence analysis of a new species from china, Grateloupia boaoensis Wang et Luan sp.(Halymeniaceae, Rhodophyta). Acta Hydrobiologca Sinica, 38 (5) : 938 –944. |
| Mateo-Cid L E, Mendoza-González A C, Gavio B, Fredericq S, 2005. . nov.(Halymeniaceae, Rhodophyta), a peculiar new prostrate species from tropical Pacific Mexico. Phycologia, 44 (1) : 4 –16. Doi: 10.2216/0031-8884(2005)44[4:GHSNHR]2.0.CO;2 |
| Miller K A, Hughey J R, Gabrielson P W, 2009. Research note:first report of the Japanese species Grateloupia lanceolata(Halymeniaceae, Rhodophyta) from California, USA. Phycological Research, 57 (3) : 238 –241. Doi: 10.1111/j.1440-1835.2009.00542.x |
| Moncalvo J M, Lutzoni F M, Rehner S A, Johnson J, Vilgalys R, 2000. Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Systematic Biology, 49 (2) : 278 –305. Doi: 10.1080/10635159950173852 |
| Okamura K, 1936. Japanese Algae. Uchida-rokakuho, Tokyo : 964p . |
| Posada D, Buckley T R, 2004. Model selection and model averaging in phylogenetics:advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology, 53 (5) : 793 –808. Doi: 10.1080/10635150490522304 |
| Ronquist F, Huelsenbeck J P, 2003. MrBayes 3:bayesian phylogenetic inference under mixed models. Bioinformatics, 19 (12) : 1 572 –1 574. Doi: 10.1093/bioinformatics/btg180 |
| Sheng Y W, Zhang W, Zhao D, Wang H W, 2012. A morphological and molecular assessment of the genus Sinotubimorpha(Halymeniaceae, Rhodophyta). Journal of Systematics and Evolution, 50 (2) : 146 –152. Doi: 10.1111/jse.2012.50.issue-2 |
| Swofford D L, 2003. PAUP*:Phylogenetic Analysis Using Parsimony(* and Other Methods). Version 4.0 beta 10. Sinauer Associates, Sunderland, Massachusetts . |
| Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, 2011. MEGA5:molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28 (10) : 2 731 –2 739. Doi: 10.1093/molbev/msr121 |
| Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G, 1997. The CLUSTAL-X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25 (24) : 4 876 –4 882. Doi: 10.1093/nar/25.24.4876 |
| Wang H W, Guan Y, Zhao F Q, Zhao D, 2015. Grateloupia huanghaiensis sp. nov.(Halymeniaceae, Rhodophyta), a peculiar new species from China. Marine Biology Research, 11 (4) : 396 –404. Doi: 10.1080/17451000.2014.932913 |
| Wang H W, Kawaguchi S, Horiguchi T, Masuda M, 2000. Reinstatement of Grateloupia catenata(Rhodophyta, Halymeniaceae) on the basis of morphology and rbcL sequences. Phycologia, 39 (3) : 228 –237. Doi: 10.2216/i0031-8884-39-3-228.1 |
| Wang H W, Kawaguchi S, Horiguchi T, Masuda M, 2001. A morphological and molecular assessment of the genus Prionitis J. Agardh(Halymeniaceae, Rhodophyta). Phycol ogical Res earch, 49 (3) : 251 –261. Doi: 10.1111/pre.2001.49.issue-3 |
| Wilkes R J, Mcivor L M, Guiry M D, 2005. Using rbcL sequence data to reassess the taxonomic position of some Grateloupia and Dermocorynus species(Halymeniaceae, Rhodophyta) from the north-eastern Atlantic. Eur opean J ournal of Phycol ogy, 40 (1) : 53 –60. |
| Xia B M, 2004. Flora Algarum Marinarum Sinicarum(Tomus Ⅱ) Rhodophyta(No. Ⅲ) Gelidiales Cryptonemiales Hildenbrandiales. Science Press, Beijing, China. p : 139 –140. |
| Yendo K, 1920. Novae algae Japoniace. Bot. Mag., 34 : 1 –12. |
| Yu L, Wang H W, Luan R X, 2013. . nov.(Halymeniaceae, Rhodophyta):a new species from south China sea based on morphological observation and rbcL gene sequences analysis. BioMed Research International, 2013 : 9 . |
| Zhang W, Wang H W, Zhao D, Sheng Y W, Luan R X, 2012. Morphological observation and rbcL gene sequence analysis of Grateloupia orientalis(Rhodophyta), a new record of the Hainan province. Marine Sciences, 36 (7) : 109 –116. |
| Zhao D, Wang H W, Sheng Y W, Lü J Z, Luan R X, 2012. Morphological observation and rbcL gene sequences studies of two new species, Grateloupia dalianensis H H.W. Wang et D. Zhao, sp. nov. and G. yinggehaiensis H. W.Wang et R. X. Luan, sp. nov.(Halymeniaceae, Rhodophyta) from China. Acta Oceanol ogica Sini ca, 31 (2) : 109 –120. Doi: 10.1007/s13131-012-0197-9 |
 2016, Vol. 34
2016, Vol. 34