Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LIU Jiao(刘娇), LI Xianchao(李先超), TANG Xuexi(唐学玺), ZHOU Bin(周斌)
- Identification and characterization of a DnaJ gene from red alga Pyropia yezoensis(Bangiales, Rhodophyta)
- Journal of Oceanology and Limnology, 34(2): 405-411
- http://dx.doi.org/10.1007/s00343-015-4321-5
Article History
- Received Nov. 17, 2014
- accepted in principle Feb. 14, 2016
Members of the DnaJ family (also known as heat shock protein 40 kDa, HSP40) are chaperone proteins that play pivotal roles in various cellular processes, such as protein folding, protein transporting and the cellular responses to stress (Walter and Buchner, 2002). The main function of DnaJ proteins is protein folding (Langer et al., 1992). After proteins or polypeptides are produced, they must be folded into their correct 3-dimensional structure to perform their function. Proteins that aid such 3-dimensional folding are termed chaperones. Chaperones bind to unfolded or partially folded proteins, prevent misfolding and assembly or reassembly of protein structures (Sung et al., 2001). DnaJ is also known as a co-chaperone because it assists another type of chaperone (DnaK) during protein folding (Rüdiger et al., 2001). Such proteins are ubiquitous and present in diverse organisms, including archaea, bacteria and eukaryotes (Kültz, 2005). The structure and regulation mechanism of DnaJ genes and proteins have been intensively studied in mammals, plants and bacteria. For example, six DnaJ homologs have been identified in Escherichia coli and 22 in Saccharomyces cerevisiae, and genomewide analysis revealed 41 DnaJ/Hsp40 family members in humans (Qiu et al., 2006). However, little information on DnaJ proteins from marine red algae is available.
Pyropia (also known as Porphyra) is a marine red algae genus, among which several species are economically important (Buschmann et al., 2001). Pyropia develops a dimorphic life cycle comprising a filamentous sporophyte and a leafy gametophyte phase (Wang et al., 2007). The developmental aspects of its life cycle have been intensively investigated (Fan et al., 2008). A developed laboratory culture method for this genus has made it possible to reproduce its complete life cycle artificially within several months. Using this method, Pyropia has recently been adopted as a model organism for fundamental and applied research in marine red algae (Israel, 2010). Furthermore, its genome size was estimated as 2.6– 5.3×108 bp and comprises 2–7 chromosomes, which is similar to those of Arabidopsis thaliana and Oryza sativa, which offers many opportunities to investigate the genetics and genomics of Pyropia (Kitade et al., 1998). Among this genus, Pyropia yezoensis M. S. Hwang et H. G. Choi (also known as Porphyra yezoensis Ueda) is one of the most investigated species because of its commercial importance (Behura et al., 2002). Expressed sequence tags (ESTs) have been collected for various species and large numbers of ESTs from P . yezoensis and Pyropia haitanensis have been deposited in public databases (Lee et al., 2000; Nikaido et al., 2000; Asamizu et al., 2003; Fang et al., 2007), which makes it possible to clone and analyze the DnaJ gene from P . yezoensis .
Our research describes the molecular characterization of the full-length cDNA sequence encoding a DnaJ protein (designated as Py DnaJ) from P . yezoensis . The main objects of our study were: 1) to clone and identify the full-length cDNA sequence of Py DnaJ; 2) to investigate the mRNA expression profile of Py DnaJ during its dimorphic life cycle; and 3) to estimate the mRNA expression profile of Py DnaJ during desiccation stress.
2 MATERIAL AND METHOD 2.1 Algae collection and culture conditionThe filamentous sporophytes of P . yezoensis were purchased from the Pyropia Culture Collection Center, Marine Fisheries Research Institute of Jiangsu Province and were maintained in Provasoli’s enriched seawater (PES) liquid medium in glass aquaria at approximately 45 μmol/(m 2 ∙s)(light (L):dark (D) cycle, 12 h:12 h) at 18–20 ℃. Leafy gametophytes of the red algae were collected in July 2012 from the Taipingjiao intertidal in Qingdao, Shandong Province, China. The material was stored in plastic boxes without seawater and transported to the laboratory on ice and also maintained in glass aquaria containing PES liquid medium at about 45 μmol/(m 2 ∙s)(L:D cycle 12 h:12 h) at 8–10 ℃.
2.2 Total RNA extraction and cDNA synthesisAbout 50 mg of sporophytic red algae material was used for total RNA isolation, which used an MagExtractor™ RNA extraction kit (Toyobo, Kitaku, Osaka, Japan). Total raw RNA was treated with 0.1 U/μL RNase-free DNase I RQ1 (Promega, Madison, WI, USA) at 37℃ for 30 min. After extraction by phenol:chloroform:isoamylalcohol (50:49:1) and precipitation with ethanol, the total purified RNA was reverse transcribed using an oligo dT -adaptor primer (5 ' -GGATCCGAATTCCCCGGG (T)24 -3 ') using Superscript™ III Reverse Transcriptase (Life Technologies, Carlsbad, CA, USA) for 1 h at 55℃. The obtained total cDNA sample was employed to amplify the cDNA sequence of Py DnaJ.
2.3 Cloning of the full-length cDNA of PyDnaJAt the time of the experiment, 22 436 ESTs from P . yezoensis were available in the public database dbEST at NCBI. Among them, one 487 bps sequence (GenBank accession No. AV430823) was homologous to the 5 ' region of Viola baoshanensis DnaJ. This EST was selected to clone the full-length cDNA sequence of Py DnaJ. The 5 ' end of the Py DnaJ was already completed in the known EST sequence. Two gene specific primers, Py DnaJ_Race_F1(5 ' -ACTGTGAGCGTTTGTCTCCGATTGCCG- 3 ') and Py DnaJ_ Race_F2(5 ' -GCATTCGCAGTGTCGGTCCCTCTTGAC-3 '), were used to clone the 3 ' end of the Py DnaJ cDNA using the 3 ' -rapid identification of cDNA ends (RACE) technique. The obtained PCR products were ligated into the T-A cloning vector pMD19-T simple (TaKaRa, Otsu, Shiga, Japan) and transformed into E . coli DH5α competent cells (TransGen, Beijing, China). The obtained recombinants were identified through blue-white selection on Luria-Bertani (LB) plates containing ampicillin, and white colonies were subsequently screened using PCR with BcaBEST Sequencing Primers M13-47(5 ' -CGCCAGGGTTTTCCCAGTCACGAC- 3 ') and RV-M (5 ' -GAGCGGATAACAATTTCACACAGG- 3 ')(TaKaRa, Otsu, Shiga, Japan). Positive clones were sequenced using an ABI Prism™ 3730 automated DNA sequencer (Thermo Fisher Scientific, Carlsbad, CA, USA) to verify the full-length cDNA sequence.
2.4 Sequence feature analysisThe full-length cDNA and deduced polypeptide sequence of Py DnaJ were analyzed using the BLAST software (http://www.ncbi.nlm.nih.gov/blast). Signal peptides were predicted using the SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP). Functional domains and motifs were predicted using the normal mode of the Simple Modular Architecture Research Tool (SMART 7.0)(http://smart.embl-heidelberg.de).
2.5 Multiple sequences alignment and phylogeneticanalysis The amino acid sequences of DnaJ homologs used for phylogenetic analysis were from P . yezoensis (AFJ54624; this study), P . haitanensis (AIS25018), Galdieria sulphuraria (XP_005707432 and XP_005707879), A . thaliana (AAB49030), O . sativa (AAX95135), Homo sapiens (BAA02656), Rattus norvegicus (NP_001020582), Drosophila melanogaster (NP_650283) and Mus musculus (NP_032324). The multiple alignment was performed using CLUSTALW (www.ch.embnet.org/software/ ClustalW.html) and the multiple alignment show program (www.bioinformatics.org/sms/multi_align. html). An unrooted phylogenetic tree was constructed based on the deduced amino acid sequences of Py DnaJ and other DnaJs using the seaview4 software (http://doua.prabi.fr/software/seaview). To derive the confidence value for the phylogeny analysis, bootstrap trials were replicated 1 000 times.
2.6 Desiccated leafy gametophyte samples preparationThe desiccation treated leafy gametophytes of P . yezoensis were prepared according to a previous report (Zou and Gao, 2002). About 100 mg of leafy red algae were desiccated in an incubator at 45 μmol/(m 2 ∙s), 20% relative humidity and 10℃. A series of desiccation levels (0%, 15%, 30%, 45% and 60%) was obtained by varying the duration of exposure. Desiccation degree was expressed as the percentage loss of water from the desiccated samples. The water lost (WL, %) was calculated using the formula: WL =(W0 – Wt)/(W0 – Wd)×100%, where W0 stands for the initial wet weight measured after removing surface water drops with tissue paper, Wt stands for the desiccated weight after a certain time interval and Wd stands for the dry weight (after incubation at 85℃ for 24 h).
2.7 Real-time PCR detection of Py DnaJ mRNAexpression Total raw RNA was extracted from sporophytes, and normal and desiccated gametophytes of P . yezoensis . cDNA was synthesized as described in Section 2.2 and diluted to approximately 20 ng/μL. The relative mRNA expression levels of Py DnaJ in different phases of life cycle and in different desiccation states were detected by real-time PCR using the real-time PCR Master Mix (Toyobo, Kitaku, Osaka, Japan). The real-time PCR assay was performed on a Bio-Rad iQ5™ Multicolor Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA) and carried out in a total volume of 25.0 μL, containing 1×SYBR real-time PCR Master Mix, 4.0 μL of the diluted cDNA mix, 0.4 mmol/L of each primer and 7.5 μL of DEPC-treated RNA-free water. Two Py DnaJ gene specific primers, Py DnaJ_qRT_F (5 ' -GCTTCAAAGTCAATTTCCCGACCA-3 ') and Py DnaJ_qRT_R (5 ' -CACTCTTCAACCTCAAAGCCATCC- 3 '), were used to amplify a product of 118 bps from the cDNA template. The obtained PCR product was sequenced to verify the specificity of the real-time PCR assay. 18S rRNA (SSU) was used as an internal control. Two 18S gene specific primers, Py 18S_real-time_F (5 ' -CGACCGTTTACTGTGAAG- 3 ') and Py 18S_real-time_R (5 ' -GACAATGAAATACGAATGCC- 3 '), were used to amplify a 160-bp fragment. The thermal profile for the real-time PCR assay was 94℃ for 3 min; followed by 40 cycles of 94℃ for 15 s, 60℃ for 40 s, 72℃ for 30 s. A dissociation curve assay of the amplification products was performed at the end of the real-time PCR reaction to confirm that only one specific product was amplified and detected. After the real-time PCR program, the data were analyzed using the Bio-Rad optical system software using the 2 -ΔΔ C t method (Livak and Schmittgen, 2001), according to the previous reports (Wang et al., 2011; Li et al., 2012). All data are shown as the relative abundance of mRNA expression level (mean±S.E.(n =5)). The data generated from real-time PCR assay were subjected to a one-way analysis of variance (one-way ANOVA), followed by an unpaired, two-tailed Student’s t -test. Differences were considered significant at P<0.05.
3 RESULT 3.1 Cloning the full-length cDNA sequence of Py DnaJ geneA fragment of 1 607 bps at the 3 ' end of Py DnaJ cDNA was obtained by 3 ' RACE based on the 487 bps EST sequence AV430823. A 1 956-bp nucleotide sequence representing the full-length cDNA sequence of Py DnaJ gene was deposited in GenBank under the accession No. JQ793376. The complete nucleotide sequence and the deduced amino acids of Py DnaJ gene are shown in Fig. 1. The complete sequence of Py DnaJ cDNA consisted of a 497-bp 5 ' untranslated region (UTR), a 169-bp 3 ' UTR and a 1 290-bp open reading frame (ORF). The 3 ' UTR contained a polyA tail but no putative polyadenylation signal (AATAAA). The ORF encoded a polypeptide of 429 amino acid residues with a calculated molecular mass of about 46 kDa and a theoretical isoelectric point of 6.884. No signal peptide in the deduced amino acid sequence was revealed by SignalP program analysis. A DnaJ domain (T 33 to K 88), a DnaJ_zf domain (G 150 to K 230) and a DnaJ_C (G 267 to A 369) domain were also predicted.
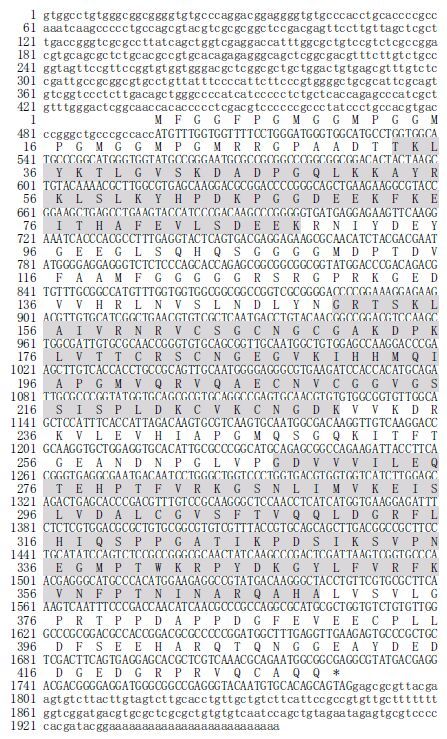
|
| Figure 1 Nucleotide and deduced amino acid sequences of Py DnaJ (GenBank accession number JQ793376) The number of nucleotides and deduced amino acid residues are shown in the left margin. The DnaJ domain (T 33 to K 88), DnaJ_zf domain (G 150 to K 230) and DnaJ_C (G 267 to A 369) domain are shaded. |
The deduced amino acid sequence of Py DnaJ showed high similarity with other reported DnaJs, such as 96% identity with that of P . haitanensis, 50% and 55% with G . sulphuraria (XP_005707432 and XP_005707879), and 48% with O . sativa . An alignment of the amino acid sequence of Py DnaJ with that of P . haitanensis HSP40/DnaJ is shown in Fig. 2. The phylogenetic tree based on amino acid sequences from multiple DnaJs proteins showed three main branches. As expected, Py DnaJ was clustered with the DnaJs from P . haitanensis and G . sulphuraria (Fig. 3).
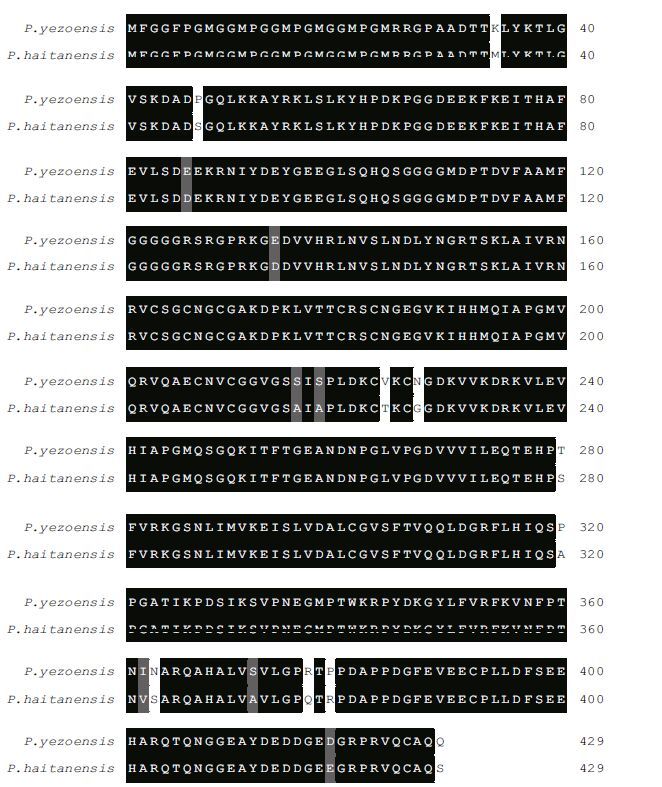
|
| Figure 2 Fig.2 Multiple alignment of PyDnaJ with P. haitanensis HSP40/DnaJ |
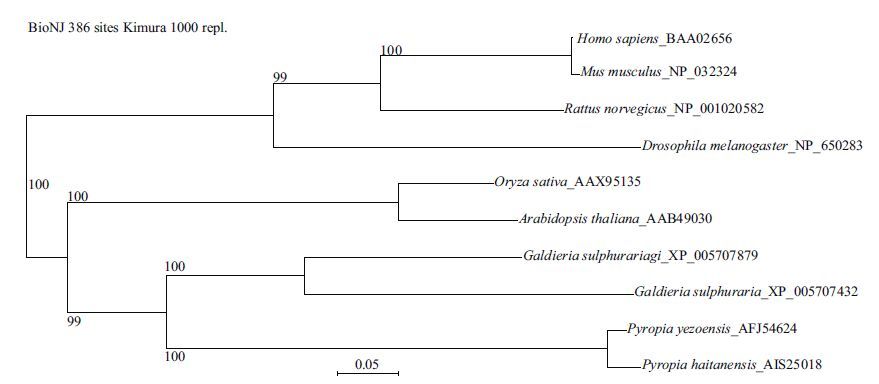
|
| Figure 3 Consensus neighbor-joining tree based on the sequences of DnaJs from different organisms The numbers at the forks indicated the bootstrap values. |
The relative mRNA expression level of Py DnaJ was detected in both sporophyte and gametophyte phases of P . yezoensis (Fig. 4). The relative mRNA expression level in sporophytes was higher (by approximately 14.3-fold) than that in gametophytes (P<0.05).

|
| Figure 4 The mRNA expression levels of Py DnaJ gene detected via real-time quantitative PCR at different life cycle stages of P . yezoensis The mRNA expression level of the gametophyte was set to 1.0. |
During the desiccation process, the relative mRNA expression level of Py DnaJ was significantly upregulated after 15% water loss, and then increased to a maximum level at 60% water loss, which was 5.1-fold above the non-desiccated level (P<0.05)(Fig. 5).
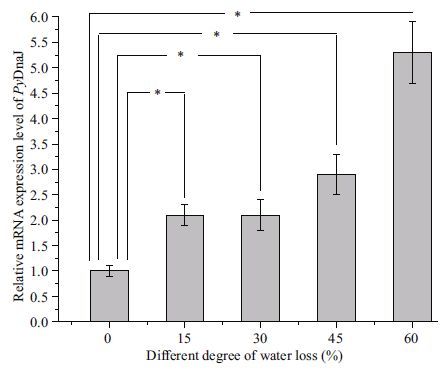
|
| Figure 5 Real-time quantitative PCR analysis the mRNA expression level of Py DnaJ gene in P . yezoensis gametophytes during desiccation 0%, 15%, 30, 45% and 60% represent the degree of water loss. The mRNA expression level at 0% desiccation was set to 1.0. |
The structure and regulation mechanism of DnaJ genes and proteins has been studied intensively in mammals (Qiu et al., 2006), plants and bacteria. However, little information of DnaJ proteins from marine red algae is available. Here, we isolated the full length cDNA for P . yezoensis DnaJ (Py DnaJ). Typical function domains were found in the deduced amino acid sequence of Py DnaJ, including a DnaJ domain, a DnaJ_zf domain and a DnaJ_C domain (Ohtsuka and Hata, 2000). The full-length DnaJ cDNA sequence from P . yezoensis shared high similarity with other DnaJ protein sequences. In addition, there were few differences between Py DnaJ and P . haitanensis HSP40/DnaJ. In the phylogenetic tree based on amino acid sequences from multiple DnaJs proteins, Py DnaJ was clustered with the DnaJs from P . haitanensis and G . sulphuraria . Taken together, they suggested that Py DnaJ from P . yezoensis is an authentic member of the plant and red algae DnaJ family.
DnaJ, also known as HSP40, is involved in the heat shock response in many organisms (Feder and Hofmann, 1999). Pyropia develops a dimorphic life cycle comprising a filamentous sporophyte and a leafy gametophyte phase (Wang et al., 2007). In the present study, the relative mRNA expression level of Py DnaJ in sporophytes was significantly higher than that in gametophytes. A potential reason for this might be that the growth temperature of sporophytes was higher than that of gametophytes (Wang et al., 2007; Li et al., 2012); therefore, the sporophytes need to express more heat shock proteins, including DnaJ, in response to the higher temperature.
The main function of DnaJ is in protein folding. DnaJ proteins are also known as co-chaperones because they help DnaKs with protein folding (Langer et al., 1992). In Pyropia, the gametophyte lives in a changeable environment, where they are subjected to repeated immersion and emersion because of daily tidal fluctuations. As a result, they were exposed to a wide range of environmental stresses such as high light, osmotic stress, rapid temperature fluctuations and desiccation twice a day (Zou and Gao, 2002). Many seaweeds in intertidal areas experience extreme drying rates, reaching air dryness within hours (Kumar et al., 2011). Desiccation could trigger high drought stress, an increased release of reactive oxygen species, and an increased production of misfolded proteins (Kurepa et al., 2009; Wang et al., 2009). Our results show that the mRNA expression levels of Py DnaJ increased continuously during the whole desiccation process. Therefore, the desiccated leafy gametophyte may need more chaperones and cochaperones, including DnaJ, to refold the misfolded proteins caused by desiccation. Our results suggest that Py DnaJ plays a pivotal role in mitigating the damage to P . yezoensis caused by desiccation.
5 CONCLUSIONWe identified and characterized the full-length DnaJ cDNA from P . yezoensis . This Py DnaJ is an authentic member of DnaJ family in plants and red algae. Transcription of Py DnaJ varies in different life cycle phases and its transcription was induced during desiccation, which suggested that Py DnaJ plays a pivotal role in mitigating damage to P . yezoensis during desiccation.
6 ACKNOWLEDGMENTWe are grateful to all the laboratory members for their technical advice and helpful discussions. The authors thank the anonymous reviewers for their critical reading of the manuscript and helpful suggestions.
| Asamizu E, Nakajima M, Kitade Y, Saga N, Nakamura Y, Tabata S, 2003. Comparison of RNA expression profiles between the two generations of Porphyra yezoensis(Rhodophyta), based on expressed sequence tag frequency analysis. Journal of Phycology, 39 (5) : 923 –930. Doi: 10.1046/j.1529-8817.2003.03003.x |
| Behura S, Sahoo S, Srivastava V K, 2002. Porphyra-the economic seaweed as a new experimental system. Current Science, 83 (11) : 1313 . |
| Buschmann A H, Correa J A, Westermeier R, Del Carmen Hernández-González M, Norambuena R, 2001. Red algal farming in Chile:a review. Aquaculture, 194 (3-4) : 203 –220. Doi: 10.1016/S0044-8486(00)00518-4 |
| Fan X, Wang G C, Li D M, Xu P, Shen S D, 2008. Study on early-stage development of conchospore in Porphyra yezoensis Ueda. Aquaculture, 278 (1-4) : 143 –149. Doi: 10.1016/j.aquaculture.2008.02.037 |
| Fang X L, Fang Y J, Hu S N, Wang G C, 2007. Generation and analysis of 5318 expressed sequence tags from the filamentous sporophyte of Porphyra haitanensis(Rhodophyta). Journal of Phycology, 43 (6) : 1287 –1294. Doi: 10.1111/jpy.2007.43.issue-6 |
| Feder M E, Hofmann G E, 1999. Heat-shock proteins, molecular chaperones, and the stress response:evolutionary and ecological physiology. Annual Review of Physiology, 61 : 243 –282. Doi: 10.1146/annurev.physiol.61.1.243 |
| Israel A. 2010. The extreme environments of Porphyra, a fast growing and edible red marine macroalga. I n:Seckbach J, Chapman D J eds. Red Algae in the Genomic Age.Springer, Netherlands. p.61-75. |
| Kitade Y, Taguchi G, Shin J-A, Saga N, 1998. Porphyra monospore system(Bangiales, Rhodophyta):a model for the developmental biology of marine plants. Phycological Research, 46 (1) : 17 –20. Doi: 10.1111/pre.1998.46.issue-1 |
| Kültz D, 2005. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol., 67 : 225 –257. Doi: 10.1146/annurev.physiol.67.040403.103635 |
| Kumar M, Gupta V, Trivedi N, Kumari P, Bijo A J, Reddy C R K, Jha B, 2011. Desiccation induced oxidative stress and its biochemical responses in intertidal red alga Gracilaria corticata(Gracilariales, Rhodophyta). Environmental and Experimental Botany, 72 (2) : 194 –201. Doi: 10.1016/j.envexpbot.2011.03.007 |
| Kurepa J, Wang S H, Li Y, Smalle J, 2009. Proteasome regulation, plant growth and stress tolerance. Plant Signaling & Behavior, 4 (10) : 924 –927. |
| Langer T, Lu C, Echols H, Flanagan J, Hayer M K, Hartl F U, 1992. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature, 356 (6371) : 683 –689. Doi: 10.1038/356683a0 |
| Lee E K, Seo S B, Kim T H, Sung S K, An G, Lee C H, Kim Y J, 2000. Analysis of expressed sequence tags of Porphyra yezoensis. Molecules and Cells, 10 (3) : 338 –342. |
| Li X C, Xing Y Z, Jiang X, Qiao J, Tan H L, Tian Y, Zhou B, 2012. Identification and characterization of the catalase gene Py CAT from the red alga P yropia yezoensis(Bangiales, Rhodophyta). Journal of Phycology, 48 (3) : 664 –669. Doi: 10.1111/j.1529-8817.2012.01152.x |
| Livak K J, Schmittgen T D, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. M ethods, 25 (4) : 402 –408. |
| Nikaido I, Asamizu E, Nakajima M, Nakamura Y, Saga N, Tabata S, 2000. Generation of 10, 154 expressed sequence tags from a leafy gametophyte of a marine red alga, Porphyra yezoensis. DNA Research, 7 (3) : 223 –227. Doi: 10.1093/dnares/7.3.223 |
| Ohtsuka K, Hata M, 2000. Molecular chaperone function of mammalian Hsp70 and Hsp40-a review. International Journal of Hyperthermia, 16 (3) : 231 –245. Doi: 10.1080/026567300285259 |
| Qiu X B, Shao Y M, Miao S, Wang L, 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cellular and Molecular Life Sciences, 63 (22) : 2560 –2570. Doi: 10.1007/s00018-006-6192-6 |
| Rüdiger S, Schneider-Mergener J, Bukau B, 2001. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. The EMBO Journal, 20 (5) : 1042 –1050. Doi: 10.1093/emboj/20.5.1042 |
| Sung D-Y, Kaplan F, Guy C L, 2001. Plant Hsp70 molecular chaperones:protein structure, gene family, expression and function. Physiologia Plantarum, 113 (4) : 443 –451. Doi: 10.1034/j.1399-3054.2001.1130402.x |
| Walter S, Buchner J, 2002. Molecular chaperones-cellular machines for protein folding. Angewandte Chemie International Edition, 41 (7) : 1098 –1113. Doi: 10.1002/(ISSN)1521-3773 |
| Wang M Q, Hu J J, Zhuang Y Y, Zhang L, Liu W, Mao Y X, 2007. In silico screening for microsatellite markers from expressed sequence tags of Porphyra yezoensis(Bangiales, Rhodophyta). Journal of Ocean University of China, 6 (2) : 161 –166. Doi: 10.1007/s11802-007-0161-z |
| Wang M Q, Mao Y X, Zhuang Y Y, Kong F N, Sui Z H, 2009. Cloning and analysis of calmodulin gene from Porphyra yezoensis Ueda(Bangiales, Rhodophyta). Journal of Ocean University of China, 8 (3) : 247 –253. Doi: 10.1007/s11802-009-0247-x |
| Wang M Q, Yang J L, Zhou Z, Qiu L M, Wang L L, Zhang H, Gao Y, Wang X Q, Zhang L, Zhao J M, Song L S, 2011. A primitive Toll-like receptor signaling pathway in mollusk Zhikong scallop Chlamys farreri. Developmental and Comparative Immunology, 35 (4) : 511 –520. Doi: 10.1016/j.dci.2010.12.005 |
| Zou D H, Gao K S, 2002. Effects of desiccation and CO2 concentrations on emersed photosynthesis in Porphyra haitanensis(Bangiales, Rhodophyta), a species farmed in China. European Journal of Phycology, 37 (4) : 587 –592. Doi: 10.1017/S0967026202003876 |
 2016, Vol. 34
2016, Vol. 34


