Institute of Oceanology, Chinese Academy of Sciences
Article Information
- Ming XUE(薛明), Huafang LIANG(梁华芳), Yaoyao HE(何瑶瑶), Chongqing WEN(温崇庆)
- Characterization and in-vivo evaluation of potential probiotics of the bacterial flora within the water column of a healthy shrimp larviculture system
- Journal of Oceanology and Limnology, 34(3): 484-491
- http://dx.doi.org/10.1007/s00343-016-5024-2
Article History
- Received: Jan. 21, 2015
- Accepted: Mar. 27, 2015
With the increasing requirements for shrimp products worldwide,the production of penaeid shrimp,including the reputable species,Litopenaeus vannamei,urgently needs to be largely improved. Thus far,the lifecycle of cultured shrimp is mainly subjected to intensive aquaculture conditions,which are characterized by high stocking density,high inputs,and low outputs. For the vulnerable shrimp larvae,this intensive mass nursing environment could easily lead to microdysbiosis and subsequent outbreaks of disease,or even mass larval mortality (Xu et al.,1999; FAO,2003) . Moreover,bacterial infection caused by opportunistic pathogens is frequently reported in shrimp larviculture,particularly with respect to vibriosis (Vandenberghe et al.,1999; Xu et al.,1999; Soto-Rodríguez et al.,2006) .
On the other hand,neutral and beneficial bacteria in aquatic ecosystems are recognized as important drivers of biogeochemical processes,not only in nutrient and energy flow,but also in disease control,water quality,and as potential feed (Blancheton et al.,2013) . Relevant studies associated with marine larviculture systems have been conducted to characterize the bacterial communities (Schulze et al.,2006; Sakami et al.,2014) . However,the composition and dynamic of the bacterial community in rearing water of the L. vannamei larviculture system is still unavailable,although Vandenberghe et al. (1999) reported certain vibrios associated with L. vannamei larvae,postlarvae,and hatchery probionts. Garcia and Olmos (2007) reported that bacteria with high G+C content and γ-proteobacteria predominated in L. vannamei larvae. Additionally,Pangastuti et al. (2010) reported the bacterial communities associated with egg,newly hatched nauplii,and 24 and 48 h old nauplii of L. vannamei.
In addition,isolation of probiotic bacteria from the aquatic environment where they grow optimally is a better approach as strain characteristics are dependent upon the surrounding water they thrive in (Schulze et al.,2006) . Thus,a systematic insight into the bacterial community of a healthy shrimp hatchery is essential. This study was designed firstly to characterize the composition and dynamics of the bacterial community in rearing water of L. vannamei larvae. The probiotic potential of bacterial isolates on larvae was then investigated.
2 MATERIAL AND METHOD 2.1 Rearing of shrimp larvaeOne of the larviculture ponds at a commercial shrimp hatchery in Zhanjiang,China was selected for the study. Prior to larvae stocking,the indoor cement pond (5 m 4 m 1.5 m) was filled with filtered seawater through a 300-mesh gauze with a final volume of 20 m3. Seawater was disinfected with 20 mL/m3 of formaldehyde and aerated vigorously for 24 h. L. vannamei larvae at stage nauplius 3 (N3) ,obtained from the specific-pathogen-free broodstock,were transferred to the pond with a stocking density of 200 larvae per litre. The rearing water was aerated constantly,and the temperature,salinity,and pH were maintained in ranges of 30.6-32.8°C,30.7-32.2,and 7.88-8.13,respectively.
Commercial spirulina powder and shrimp flakes were used to feed larvae beginning at late N6 stage and six times daily. When larvae developed to late zoea 3 (Z3) ,spirulina powder was stopped,Artemia nauplii were added four times daily. Throughout the 10-day larval rearing period,no water was exchanged,no antibiotic or commercial probiotics was supplemented,and no disease symptoms appeared by daily observation. Additionally,the larvae at postlarvae 1 (PL1) stage presented a high survival rate of 70.5%.
2.2 Sample collectionWater samples were collected at eight time points,which corresponding to the larval stages N6,Z1,Z2,Z3,Mysis 1 (M1) ,M2,M3,and PL1 at 1,2,3,4,5,6,8,and 10 days,respectively after the onset of stocking N3 larvae. At each time point,2 L of water samples,corresponding to four aliquots of 500 mL from four locations in the larviculture pond,were sampled by a handled sterile beaker at approximately 10 cm below the water surface and loaded in aseptic polyethylene bottles. For each sample,1 L of water was passed through a 0.22-μm filter. The filters were stored at -20°C until DNA extraction.
2.3 Bacterial isolation and 16S rRNA gene sequencingWater sample of each larval stage was tenfold diluted serially and appropriate dilutions were spread- plated on marine 2216E agar and incubated at 28°C for 5 d. For each sample,about 40 well-isolated colonies were randomly selected and divided into differentiated morphotypes according to colony characteristics. Then a total of 181 representative colonies were picked up and re-streaked to obtain pure cultures.
The genomic DNA of each isolate was extracted using a bacterial genomic extraction kit following the manufacturer’s instructions (Omega,Norcross,GA,USA) . The universal primers 27F and 1492R were used to amplify the 16S rRNA genes of the isolates as previously described (Wen et al.,2009) and the amplified products were first sequenced with primer 27F. If more than 99.5% sequence similarity were observed among the same morphotype isolates from different samples,then only one isolate was selected as a representative strain for further sequencing with primer 1492R and assembled to obtain the almost full-length 16S rRNA gene sequence. The sequencing was performed by Shanghai Sangon Co.,Ltd. with the ABI 377 DNA sequencer (Applied Biosystems,Carlsbad,CA,USA) .
2.4 PCR-DGGE analysis of water samplesWater DNA samples were extracted from the filters with Water DNA Kit (Omega) . PCR amplification and DGGE analysis of the V3-V5 region of the 16S rRNA genes,excision,and re-amplification of the dominant bands were performed as described by Jiang et al. (2011) . The re-amplified products were ligated into the pMD20-T vector (TaKaRa,Dalian,China) and transformed into Escherichia coli DH5α competent cells. Positive clones were selected for sequencing using M13 primers. After removing the primers region,the taxonomic identification of the sequences was performed as described below.
2.5 Taxonomic identification and phylogenetic analysisThe 16S rRNA gene sequences of the representative strains and DGGE bands have been deposited in GenBank under accession numbers KP301091 to KP301135. They were compared with available 16S rRNA gene sequences from GenBank via the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/,Kim et al.,2012) to get taxonomic information.
The sequences were aligned by ClustalW program together with the most similar sequences,especially the sequences of type strains. Additionally,a neighbor- joining phylogenetic tree was constructed with Kimura 2-parameter model using 1 000 bootstrap replicates with the MEGA5 software (Tamura et al.,2011) .
2.6 In-vivo larval experiments of bacterial isolatesTo select the potentially probiotic bacteria,hemolysis was firstly analyzed on blood agar plates for all the 27 representative strains. Hemolytic negative strains were then individually evaluated in in-vivo experiments with shrimp larvae. All treatments and control were performed in quadruplicates. Each strain proliferated in marine 2216E medium and logarithmic phase cells were harvested by centrifugation. The collected pellets were rinsed three times with sterile seawater,resuspended in seawater,and spectrophotometrically adjusted to a concentration of 1012 colony-forming units (CFU) /L for use. The N3 larvae were transferred to 16-L plastic buckets containing 12 L of aerated seawater with initial stocking density of 200 larvae/L. Twelve milliliters of cell suspension of each strain was added to the bucket to achieve a final concentration of 109 CFU/L in water.developed to Z3 stage. During the 4-day experimental period,the larvae were fed as described above and no water was exchanged. Water temperature,salinity,and pH were maintained in ranges of 30.3-32.5°C,29.5-30.7,and 7.85-8.09,respectively.
2.7 Statistical analysisThe percentages of larval survival were transformed to the arcsine of the square root for statistical analysis with the software SPSS 16.0 for Windows (SPSS Inc.,Chicago,IL,USA) . Dunnett t-test was used to detect significance (P<0.05) of survival rates between treatments and the control.
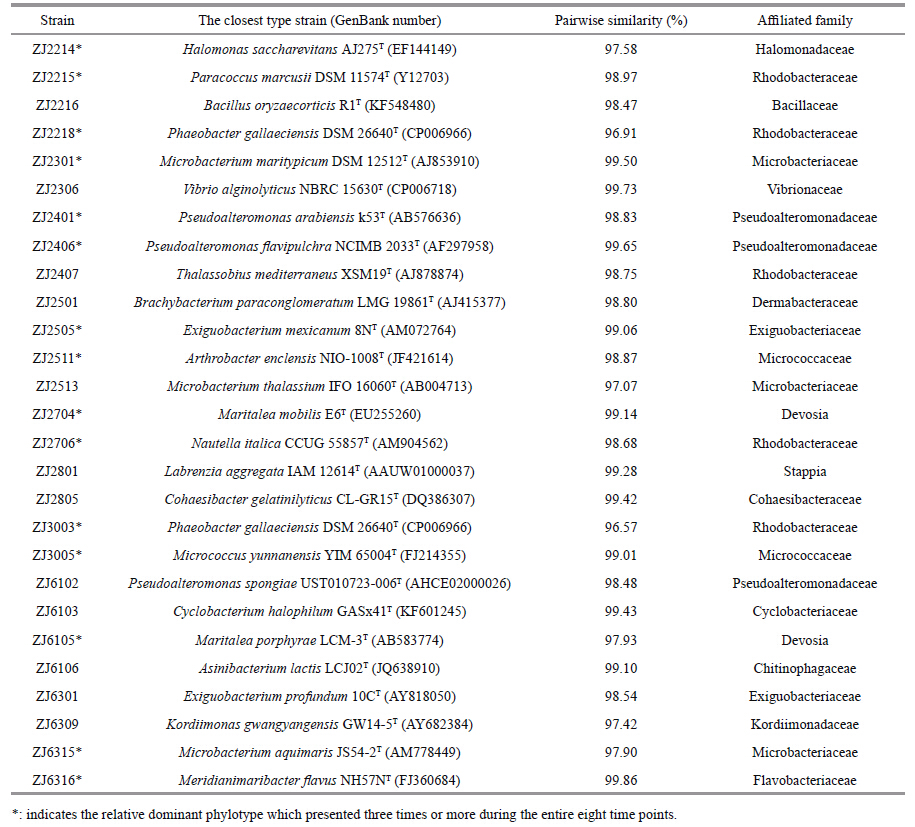
3 RESULT 3.1 Culture-based analysis of bacterial floraA total of 27 phylotypes,ranged from 10 to 14 phylotypes for each of the eight time points,were identified at the species level by EzTaxon analysis of 16S rRNA gene sequences. The most similar type strains to the representative isolates are shown in Table 1. These 27 phylotypes were assigned into 20 genera of 16 families,which belonging to five groups of α-Proteobacteria,γ-Proteobacteria,Actinobacteria,Firmicutes,and Bacteroidetes (Fig. 1) . Among them,15 phylotypes frequently presented during the larval rearing run (Table 1) ,these relatively abundant bacteria may play key roles in shrimp larviculture system.
|

|
| Figure 1 Neighbor-joining phylogenetic tree shows the relationship between 16S rDNA sequences retrieved from the representative isolates and DGGE bands and their closest relatives |
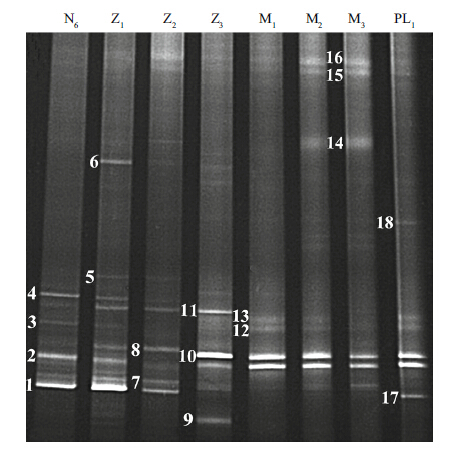
The succession of bacterial communities in larviculture water is profiled with DGGE fingerprinting of the V3-V5 region of 16S rRNA gene (Fig. 2) . The bacterial communities changed over time with significant shifts from stages zoea to mysis. A total of 18 prominent bands in the DGGE gel were excised and sequenced to confirm bacterial species composition (Table 2,Fig. 2) . Band one,with sequence affliated to family Flavobacteriaceae,appeared predominantly during the early stages from N6 to Z2,but disappeared thereafter. Bands two and seven,having 100% sequence similarities to those of the isolated strains ZJ2218 (Phaeobacter sp.) and ZJ2706 (Nautella sp.) respectively,frequently occurred at various stages,though the latter appeared comparatively weak. Band 10,with sequence ascribed to family Rhodobacteraceae,together with band two,Survival rate of larvae in each tank was recorded when 90% of surviving larvae in the control group overwhelmingly presented from stages Z3 to PL1.
|
| Figure 2 DGGE profile of bacterial community of water column associated with eight development stages of L. vannamei larvae |

|
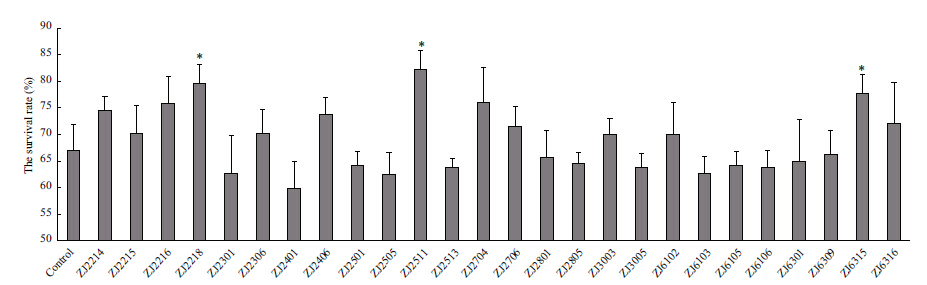
Figure 3 shows the survival rates of larvae after treated with the 26 non-hemolytic strains,individually,during stages N3 to Z3. When compared to the control,three treatments with strains ZJ2511,ZJ2218,and ZJ6315 individually showed significantly higher larval survival rates (P<0.05) . Ten other treatments,ZJ2214,ZJ2215,ZJ2216,ZJ2306,ZJ2406,ZJ2704, ZJ2706,ZJ3003,ZJ6102 and ZJ6316,also presented higher,but not significant survival rates (P>0.05) . In contrast,the remaining 13 treatments showed slightly lower larval survival than that of the control (P>0.05) .
|
| Figure 3 Survival rate of L. vannamei larvae treated without (control) and with bacterial pure cultures during stages N3 to Z3 at a concentration of 109 CFU/L |
In the present study,bacterial members within family Rhodobacteraceae under α-Proteobacteria were most frequently retrieved by either culture- based or PCR-DGGE methods. Similarly,Huang et al. (2014) reported that the bacterial community of rearing water of L. vannamei 14-day-old postlarvae was also dominated by Rhodobacteraceae. This similarity in presence of dominant bacterial group may denote strong resilience of the members of Rhodobacteraceae family to adapt to intensive aquaculture environments. Obviously,the DGGE fingerprinting differed mainly in band migration and counts for the larval stages between N6 to Z3 and M1 to PL1. This may be attributed to the shift in diet introduction,as spirulina powder was replaced by Artemia nauplii once larvae developed to late Z3. Thus the major succession in diversity and abundance of bacterial community in larval rearing water is substantially associated with food sources delivered,rather than the other two likely routes of entry of various bacteria from inlet water and broodstock (Schulze et al.,2006; Pangastuti et al.,2010) .
Furthermore,there was consistency in bacterial characterization between these two methods,i.e.,the four cultivable strains ZJ2218 (Phaeobacter sp.) ,ZJ2706 (Nautella sp.) ,ZJ3003 (Phaeobacter sp.) and ZJ6316 (Meridianimaribacter sp.) were also detected by DGGE finger-printings,which corresponded to bands 2,7,12 and 13 respectively. Sakami et al. (2014) reported also that Phaeobacter sp. and Nautella sp. were both retrieved from the dominant bands in DGGE gel of good rotifer culture,rather than growth suppression culture. Thus,the relative dominances of these bacterial floras,together with Meridianimaribacter sp.,may be indicative of their significant functions in normal larval rearing environment.
In addition,four bands,one,five,13 and 18,especially band one,markedly highlighted during the early larval stages,with sequences were all assigned to Flavobacteria-affliated species. Furthermore,isolate ZJ6316 presented the same sequence as that of band 13 and both prevailed during the latter larval phase (Table 1 and Fig. 1) . This abundance of Flavobacteria species could contribute to the degradation of multiple organic matters in eutrophication water of shrimp hatchery pond (Beardsley et al.,2011) .
In the in-vivo experiments,13 out of the 26 non- hemolytic strains conferred shrimp larvae higher survival when applied as water additives,this may indicate that these bacteria are at least not pathogenic to shrimp larvae and are good candidates for further probiotics testing. Moreover,these favorable strains were repeatedly isolated from different larval stages. Hence,they may regulate system processes crucial for larvae health. Notably,sequences of strains ZJ2218 and ZJ3003,together with sequences from bands two,12,and 14,were affliated with the genus Phaeobacter. This prevalence may be associated with significant roles of Phaeobacter for larval development and shows promise as probiotics. It is also interesting to note that Phaeobacter species have been used successfully as probiotics for other aquatic animals. Ruiz-Ponte et al. (1999) reported that marine isolate Phaeobacter gallaeciensis BS107 displayed in vitro activity against a number of bacteria,including aquaculture pathogens and benefited Pecten maximus larvae with enhanced survival. The strain Phaeobacter sp. PP-154,isolated from a bivalve hatchery,showed a broad spectrum of inhibitory activity against diverse species of Vibrio and other known aquaculture pathogens (Prado et al.,2009) . In addition,the antibacterial compound,tropodithietic acid,produced by these bacteria likely contributes to their probiotic effects (D’Alvise et al.,2012) .
Recently,Fu et al. (2014) reviewed that Arthrobacter,a genus of Gram-positive bacteria,was able to degrade toxic organic compounds,remove sulphur,phosphorus,and heavy metals,and even produce a number of valuable bioactive substances. Herein,in view of the significantly higher survival of strain ZJ2511 (Arthrobacter sp.) conferred on L. vannamei larvae,detailed information for it as probiotic remains to be investigated. In shrimp aquaculture,Li et al. (2008) have reported that Arthrobacter sp. XE-7,besides its nitrification ability,could protect shrimp L. vannamei,by antagonizing the pathogenic Vibrio spp. and improving their survival and immune responses. Xia et al. (2013) found that the Arthrobacter sp. CW9 presents both probiotic and immunostimulatory properties to P. vannamei juveniles.
Remarkably,strains ZJ2401,ZJ2406,and ZJ6102 all were affliated with genus Pseudoalteromonas,which has been exclusively isolated from marine environments (Holmström et al.,2002) . Moreover,ZJ2406 and ZJ6102 strains exhibited better larval survival than the control. This is in agreement with the report of Longeon et al. (2004) that addition of Pseudoalteromonas sp. X153 to larval cultures of P. maximus induced higher survival in comparison to the control. Thus,Pseudoalteromonas shows great promises to be applied as probiotics for shrimp larvae,as they could repress the growth of pathogenic bacteria and viruses in fish and shellfish farming by synthesizing biologically active compounds with antibacterial or algicidal properties (Longeon et al.,2004) .
For phylum Actinobacteria,besides strain ZJ2511,ZJ6315 (Microbacterium sp.) also presented superior larval survival. In addition,another four isolates (ZJ2301,ZJ2513,ZJ2501,and ZJ3005) and bands 15 and 16 were ascribed to members of Actinobacteria. This may be indicative of somewhat dominance of Actinobacteria in normal shrimp larviculture system,and of importance of the Gram-positive bacteria as probiotics for aquaculture (Das et al.,2008) . Additionally,strain ZJ2306,very close to Vibrio alginolyticus,also showed better larval survival,but this positive effect remains to be determined since wide strain variation and genetic heterogeneity exist among V. alginolyticus isolates (George et al.,2005) .
The healthy L. vannamei larviculture system in this study harbored a diverse bacterial flora,which includes four phyla,Proteobacteria,Actinobacteria,Bacteroidetes,and Firmicutes. In addition,13 potentially probiotic candidates were primarily screened by enhancing larval survival when compared to the control. Among them,three favorable candidates,ZJ2218,ZJ2511,and ZJ6315,with sequences affliated with Phaeobacter sp.,Arthrobacter sp.,and Microbacterium sp.,respectively,deserve further testing regarding their significant effects on improving larval survival.
| Beardsley C, Moss S, Malfatti F, Azam F, 2011. Quantitative role of shrimp fecal bacteria in organic matter fl uxes in a recirculating shrimp aquaculture system. FEMS Microbiol. Ecol., 77 (1) : 134 –145. Doi: 10.1111/fem.2011.77.issue-1 |
| Blancheton J P, Attramadal K J K, Michaud L, Roque d'Orbcastel E, Vadstein O, 2013. Insight into bacterial population in aquaculture systems and its implication. Aquacult. Eng., 53 : 30 –39. Doi: 10.1016/j.aquaeng.2012.11.009 |
| D'Alvise P W, Lillebø S, Prol-Garcia M J, Wergeland H I, Nielsen K F, Bergh Ø, Gram L, 2012. Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS One, 7 (8) : e43996 . Doi: 10.1371/journal.pone.0043996 |
| Das S, Ward L R, Burke C, 2008. Prospects of using marine actinobacteria as probiotics in aquaculture. Appl. Microbiol. Biotechnol., 81 (3) : 419 –429. Doi: 10.1007/s00253-008-1731-8 |
| FAO. 2003. Health management and biosecurity maintenance in white shrimp(Penaeus vannamei)hatcheries in Latin America. FAO Fisheries Technical, Paper No.450. Rome. |
| Fu H L, Wei Y F, Zou Y Y, Li M Z, Wang F Y, Chen J R, Zhang L X, Liu Z H, Ding L X, 2014. Research progress on the Actinomyces arthrobacter. Adv. Microbiol., 4 (12) : 747 –753. Doi: 10.4236/aim.2014.412081 |
| Garcia A T, Olmos J S, 2007. Quantification by fluorescent in situ hybridization of bacteria associated with Litopenaeus vannamei larvae in Mexican shrimp hatchery. Aquaculture, 262 (2-4) : 211 –218. Doi: 10.1016/j.aquaculture.2006.10.039 |
| George M R, John K R, Iyappan T, Jeyaseelan M J P, 2005. Genetic heterogeneity among Vibrio alginolyticus isolated from shrimp farms by PCR fingerprinting. Lett. Appl. Microbiol., 40 (5) : 369 –372. Doi: 10.1111/lam.2005.40.issue-5 |
| Holmström C, Egan S, Franks A, McCloy S, Kjelleberg S, 2002. Anti-fouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol., 41 (1) : 47 –58. Doi: 10.1111/fem.2002.41.issue-1 |
| Huang Z B, Li X Y, Wang L P, Shao Z Z. 2014. Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquacult. Res., http://dx.doi.org/10.1111/are.12628. |
| Jiang Y, Marang L, Kleerebezem R, Muyzer G, Van Loosdrecht M C M, 2011. Eff ect of temperature and cycle length on microbial competition in PHB-producing sequencing batch reactor. ISME J., 5 (5) : 896 –907. Doi: 10.1038/ismej.2010.174 |
| Kim O S, Cho Y J, Lee K, Yoon S H, Kim M, Na H, Park S C, Jeon Y S, Lee J H, Yi H N, Won S, Chun J, 2012. Introducing EzTaxon-e:a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol., 62 (3) : 716 –721. |
| Li J Q, Tan B P, Mai K S, Ai Q H, Zhang W B, Liufu Z G, Xu W, 2008. Immune responses and resistance against Vibrio parahaemolyticus induced by probiotic bacterium Arthrobacter XE-7 in Pacific white shrimp, Litopenaeus vannamei. J. World Aquacult. Soc., 39 (4) : 477 –489. Doi: 10.1111/jwas.2008.39.issue-4 |
| Longeon A, Peduzzi J, Barthélemy M, Corre S, Nicolas J-L, Guyot M, 2004. Purification and partial identification of novel antimicrobial protein from marine bacterium Pseudoalteromonas species strain X153. Mar. Biotechnol., 6 (6) : 633 –641. Doi: 10.1007/s10126-004-3009-1 |
| Pangastuti A, Suwanto A, Lestari Y, Suhartono M T, 2010. Bacterial communities associated with white shrimp(Litopenaeus vannamei)larvae at early developmental stages. Biodiversitas, 11 (2) : 65 –68. Doi: 10.13057/biodiv |
| Prado S, Montes J, Romalde J L, Barja J L, 2009. Inhibitory activity of Phaeobacter strains against aquaculture pathogenic bacteria. Int. Microbiol., 12 (2) : 107 –114. |
| Ruiz-Ponte C, Samain J F, Sánchez J L, Nicolas J L, 1999. The benefit of a Roseobacter species on the survival of scallop larvae. Mar. Biotechnol., 1 (1) : 52 –59. Doi: 10.1007/PL00011751 |
| Sakami T, Koiso M, Sugaya T, 2014. Characterization of bacterial community composition in rotifer cultures under unexpected growth suppression. Fish Sci., 80 (4) : 757 –765. Doi: 10.1007/s12562-014-0741-y |
| Schulze A D, Alabi A O, Tattersall-Sheldrake A R, Miller K M, 2006. Bacterial diversity in a marine hatchery:balance between pathogenic and potentially probiotic bacterial strains. Aquaculture, 256 (1-4) : 50 –73. Doi: 10.1016/j.aquaculture.2006.02.008 |
| Soto-Rodríguez S A, Simoes N, Roque A, Gómez Gil B, 2006. Pathogenicity and colonization of Litopenaeus vannamei larvae by luminescent vibrios. Aquaculture, 258 (1-4) : 109 –115. Doi: 10.1016/j.aquaculture.2006.04.035 |
| Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, 2011. MEGA5:molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., 28 (10) : 2731 –2739. Doi: 10.1093/molbev/msr121 |
| Vandenberghe J, Verdonck L, Robles-Arozarena R, Rivera G, Bolland A, Balladares M, Gomez-Gil B, Calderon J, Sorgeloos P, Swings J, 1999. Vibrios associated with Litopenaeus vannamei larvae, postlarvae, broodstock, and hatchery probionts. Appl. Environ. Microbiol., 65 (6) : 2592 –2597. |
| Wen C Q, Lai X T, Xue M, Huang Y L, Li H X, Zhou S N, 2009. Molecular typing and identification of Bdellovibrio-and-like organisms isolated from seawater shrimp ponds and adjacent coastal waters. J. Appl. Microbiol., 106 (4) : 1154 –1162. Doi: 10.1111/jam.2009.106.issue-4 |
| Xia Z, Zhu M, Zhang Y, 2013. Effects of the probiotic Arthrobacter sp. CW9 on the survival and immune status of white shrimp(Penaeus vannamei). Lett. Appl. Microbiol., 58 (1) : 60 –64. |
| Xu H S, Yang X S, Li Y, 1999. Diagnosis and Control of Bacterial Diseases in Penaeid Shrimp Hatcheries. Ocean Press: Beijing, China136-152. |
 2016, 34
2016, 34


