Institute of Oceanology, Chinese Academy of Sciences
Article Information
- SU Lei(苏蕾), ZHANG Qianqian(张倩倩), GONG Jun(龚骏)
- Development and evaluation of specific PCR primers targeting the ribosomal DNA-internal transcribed spacer (ITS) region of peritrich ciliates in environmental samples
- Chinese Journal of Oceanology and Limnology, 36(3): 818-826
- http://dx.doi.org/10.1007/s00343-018-6326-3
Article History
- Received Dec. 8, 2016
- accepted in principle Feb. 3, 2017
- accepted for publication Mar. 28, 2017
2 University of Chinese Academy of Sciences, Beijing 100049, China
Free-living peritrich ciliates (Phylum Ciliophora, Class Oligohymenophorea, Subclass Peritrichia) are a highly diverse group of protozoa that are widely distributed in freshwater, brackish water, marine and terrestrial habitats (Foissner et al., 1991, 2002; Clamp and Coats, 2000; Ji et al., 2005). Peritrichs can be important grazers for bacterial and microalgal preys in the microbial food webs; they have been used as biological indicators in water quality monitoring (Sherr et al., 1988; Fried et al., 2000; Martín-Cereceda et al., 2001; Xu et al., 2009). Previous studies have demonstrated that some species (e.g., Vorticella and Carchesium spp.) may have ecologically distinct populations in different geographic locations (Gentekaki and Lynn, 2012; Sun et al., 2013). Nevertheless, how peritrich species are assembled in natural communities is not well understood.
The diversity and ecological study of peritrichs have traditionally relied on morphological identification using microscopical observations and silver staining (e.g., Ji et al., 2009), which are, however, very time-consuming in large-scale surveys. Furthermore, some species-rich taxa (e.g., Vorticella and Zoothamnium) can only be identified with subtle features, which are highly expertise-dependent (Sun et al., 2013). Recently, molecular tools have been increasingly used in microbial diversity studies, including these for peritrich ciliates (Gentekaki and Lynn, 2012; Liu and Gong, 2012; Sun et al., 2013). PCR and quantitative real-time PCR primers have been successfully designed to amplify long or short fragments of 18S rRNA genes of peritrichs from environmental samples (Liu and Gong, 2012). Compared to the 18S rDNA, the internal transcribed spacer (ITS) region of rDNA has a higher evolutionary rate, offering a better tool for the discrimination of closely related species and populations (Hillis and Dixon, 1991; Markmann and Tautz, 2005; Sun et al., 2013). However, specific primers targeting the ITS rDNA are not yet available for peritrichs in environmental samples, though the ITS regions (including two sub-regions ITS1 and ITS2) have been widely used as markers for various microbes, e.g., fungi, cyanobacteria and other ciliates (Rocap et al., 2002; Schoch et al., 2012; Stern et al., 2012; Bachy et al., 2013; Sun et al., 2013).
In this study, we aimed to design specific PCR primers to amplify the complete ITS region of freeliving peritrichs. Furthermore, we compared the ITS regions with two other potential markers (i.e., the D1 domain of the 28S rDNA and the variable region 9 (V9) of the 18S rDNA) for their relative variability among peritrichs. With these, we hope to provide new and select proper, molecular markers in addressing diversity and biogeographic issues of these ecologically important protozoa in the changing environment.
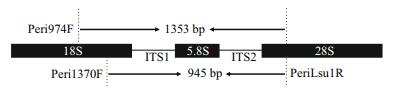
2 MATERIAL AND METHOD 2.1 Primer design and PCR testingTo design the peritrich-specific primers, the sequences containing the partial 18S rDNA, the full length of the ITS region, and the partial 28S rDNA gene were retrieved from the NCBI database, resulting in a dataset consisting of 68 peritrichs, 39 nonperitrich ciliates and 31 non-ciliate protozoans (Table S1). A forward primer Peri1370F and a reverse primer PeriLsu1R were newly designed (Table 1). These two primers target conserved regions in the 18S and the 28S rDNA, respectively (Fig. 1). We also tested the use of an existing forward primer Peri974F (Liu and Gong, 2012) with our newly designed PeriLsu1R to amplify the ITS (Table 1). The primer length, GC% and melting temperature (Tm) were calculated using Integrated DNA Technologies (http://sg.idtdna.com/site) (Table 1). The PCR products yielded by the two primer pair sets Peri974F/PeriLsu1R and Peri1370F/ PeriLsu1R were expected to cover the V9 region of 18S, the full length of the ITS regions and the D1 region of 28S, with sizes about 1 353 bp and 945 bp, respectively (Fig. 1).

|
|
| Figure 1 Schematic diagram of the rDNA region targeted by peritrich specific primers designed in this study |
The specificity of the newly designed primers was first checked in silico using probeCheck (Loy et al., 2008) and evaluated against the NCBI database using BLASTn (Altschul et al., 1997). Primer specificity was further evaluated in laboratory by taking genomic DNA extracted from peritrich isolates as the positive control and these from non-peritrich ciliates as the negative control (Table 2). DNA extraction from ciliate isolates was as previous described (Gong et al., 2013). The PCR amplifications were carried out with a Fermentas DreamtaqTM DNA Polymerase Kit (Thermo Scientific, USA). PCR programs used for the two primer pairs were as follow: 94℃ for 5 min, 35 cycles of 94℃ for 30 s, 50℃ for 1 min, 72℃ for 1.5 min, and a final extension at 72℃ for 10 min. Amplicons were checked by electrophoresis in a 1% agarose gel containing Gelview (BioTeke, Beijing, China). Band sizes were compared with a Trans2K DNA ladder (Transgen, Beijing, China).

|
The primer specificity was also examined using environmental DNA. Both surface water and sediment samples were collected from a sea cucumber farming pond in Yantai, China (37°26′44″N, 121°32′4″E) on March 20, 2013. A total of five liters of the surface water were sampled and pre-filtered through a 200-μm-pore mesh to remove most metazoan plankton and detritus. A volume of 300 mL water was then gently filtered through a 10-μm-pore polycarbonate membrane (diameter 47 mm; Millipore, Bedford, MA, USA) to collect microbial biomass. The membranes were placed immediately into 5-mL cryotubes and preserved at -80℃ for subsequent DNA extraction. Sediments were collected with a box-corer, and the top 2-cm layers were subsampled, put into plastic bags, and stored at -80℃ immediately until DNA extraction (Guo et al., 2015). The environmental DNA of both water and sediment samples were extracted separately using a FastDNA Spin Kit for Soil (MP Biomedical, USA) according to the manufacturer's instructions. The concentrations of DNA were quantified by a NanoDrop 2000C spectrophotometer (Thermo, Winmington, DE, USA).
2.3 Cloning and sequencingTwo libraries for water samples and another two for sediment samples were constructed. The PCR programs were as follows: 94℃ for 5 min, 35 cycles of 94℃ for 30 s, 55℃ (56℃ for Peri1370F/ PeriLsu1R) for 1 min, 72℃ for 1.5 min (1 min for Peri1370F/PeriLsu1R), with a final extension at 72℃ for 10 min. Quadruplicate PCR products from each DNA sample were pooled and purified using a TIANprep Midi Purification Kit (Tiangen, Beijing, China). Clone libraries were made using an InsTAclone PCR Cloning Kit (Thermo, Winmington, DE, USA). Multiple white clones were randomly picked and screened with the primers M13F and M13R. The positive clones with inserts were sequenced in both directions with an ABI 3700 sequencer (Sangon, Shanghai, China).
2.4 Sequence examination and phylogenetic analysesThe newly obtained sequences were checked using the Bellerophon to identify and remove possible chimeras (Huber et al., 2004), and OTUs were determined by Mothur v1.35.1 (Schloss et al., 2009) with a cutoff of 97%. Subsequently, the retained sequences were appended with 41 sequences retrieved from GenBank, containing ITS rDNA of peritrich species that were morphologically identified. Sequences were aligned using MAFFT (Katoh et al., 2002) and revised manually with SeaView4 (Galtier et al., 1996). The partial 18S and 28S were trimmed manually to generate an ITS alignment. The final alignment of the ITS region included 158 species and 500 sites. Both maximum likelihood (ML) and Bayesian inference (BI) algorithms were applied to build phylogenetic trees. The ML analyses were performed by RaxML v8.2 (Stamatakis, 2014), with a GTRGAMMAI model and 1 000 bootstrapping replicates. BI analyses were executed using MrBayes v. 3.2.6 (Ronquist and Huelsenbeck, 2003) with a GTR+I+G model. Four MCMC chains were run for 3 000 000 generations, with a sampling by every 100 generations. The first 25% of the generations were discarded as burn-in.
2.5 Variability of V9, ITS, and D1 region of 28S rDNA sequences among peritrichsThe fragments yielded by PCR with our new primer sets are composed of the variable region 9 (V9) of the 18S rDNA, the whole ITS (ITS1+5.8S rDNA+ITS2), and partial D1 domain of the 28S rDNA (about 350 bp at the beginning of 28S) (Fig. 1), which allowed a comparison of the relative variability of these variable regions. The whole ITS (short named as ITS), its two sub-regions ITS1 and ITS2, the 28S and the ITS+28S regions were each compared to the V9 region. After removing short sequences, the remaining 101 sequences were compiled and aligned using BioEdit (Hall, 1999). Pairwise difference was calculated by MEGA 6.0 (Tamura et al., 2013), with a Kimura 2-parameter distance model and a complete deletion. Scatter plotting, data fitting and coefficients (R2) calculation for locus pairs were performed using Microsoft Excel.
3 RESULT 3.1 Specificity of the newly designed primersThe in silico check using probeCheck and the BLAST against the NCBI database showed good specificity of the newly designed primers Peri1370F and PeriLsu1R. The primer Peri1370F matched exclusively and perfectly to peritrich sequences across most peritrich lineages, which include the families Astylozoidae, Operculariidae, Ophrydiidae, Opisthonectidae, Scyphidiidae, Vaginicolidae, Zoothaminiidae and the genera Trichodina, Campanella, Epistylis, Opisthostyla, Apocarchesium, Carchesium, Epicarchesium, Pseudovorticella and Vorticella. No matches were found for the reverse primer PeriLsu1R by probeCheck. Using the PrimerBLAST function provided by the NCBI database, only peritrich sequences were obtained in testing the two primer pairs Peri1370F/PeriLsu1R and Peri974F/ PeriLsu1R.
Using DNA from 7 peritrich isolates as template, PCR with Peri974F/PeriLsu1R and Peri1370F/ PeriLsu1R generally produced bright bands of expected sizes in the gel (Table 2). An exception was found for two Zoothamnium species and one Epicarchesium species, with which PCR did not yield positive results. Nevertheless, PCR testing with DNA of non-peritrich organisms was all negative (Table 2). PCR using the DNA extracted from water and sediment samples also yielded positive results when checked with gel electrophosis.
3.2 Classification of environmental sequences via phylogenetic analysisUsing the two newly designed primer pairs, four clone libraries were constructed for the water and sediment samples. A total of 120 clones from the 4 libraries were screened and sequenced, in which 7 sequences were probably chimeric and hence removed from subsequent analysis. Eighteen OTUs were finally determined (Fig. 2, Table S2). BLASTing the remaining 113 sequences against GenBank showed that all the best hits belonged to peritrich ciliates, with the coverage ranging of 65%–100% and the identities between 83% and 95% (Table S2). The resulted peritrich clone sequences have been submitted to GenBank, with accession numbers of KX926019–KX926131.

|
| Figure 2 A phylogenetic tree based on ITS rDNA using maximum likelihood (ML) and Bayesian inference (BI) algorithms, showing the classifications of the sequences obtained in this study (in bold) Only bootstrap values higher than 50 and 0.8 for ML and BI were given at the nodes. Dashes (-) indicate bootstrap values less than 50 or 0.8 for ML or BI single. The open and solid circles refer to the sequences that were derived from water and sediment, respectively. Letters "A" and "B" indicate clone sequences that were yield by using Peri1370F/PeriLsu1R and Peri974F/PeriLsu1R, respectively. Asterisks (*) indicate alternate topologies. |
In the phylogenetic trees (Fig. 2), the obtained environmental sequences were classified into many lineages of Peritrichia, including Pseudovorticella, clade Ⅱ of the Vorticella complex, unidentified clades of the families Vorticellidae, Vaginicoiidae and Zoothamiidae. Among these, 104 of the 113 sequences (13 of the 18 OTUs) belonged to family Vorticellidae. The amplicons belonging to family Vaginicolidae were exclusively yielded by using Peri974F/PeriLsu1R (Fig. 2); while all sequences assigned in Zoothamiidae were amplified using Peri1370F/ PeriLsu1R (Fig. 2).
Sequences from the water and sediment samples clearly separated from each other, belonging to different groups or forming distinct clusters. The sequences derived from sediment samples were assigned to Pseudovorticella and its relatives, clade Ⅱ of Vorticella, Vaginicolidae and one clade of Zoothamniidium. The sequences from surface waters fell in a cluster grouping to Epicarchesium, a clade closely related to clade Ⅱ of Vorticella, and one clade of Zoothamiidae. Only one clade affiliated to Vorticella clade Ⅱ was composed of OTUs (OTUs 8–10) from both water and sediment samples. Four clusters containing more than 3 sequences were depicted, which comprised exclusively by water or sediment sequences. The most abundant cluster (cluster 1, 59 sequences) comprised only sequences derived from water samples, whereas the clusters 2, 3 and 4 included merely sediment sequences (Fig. 2).
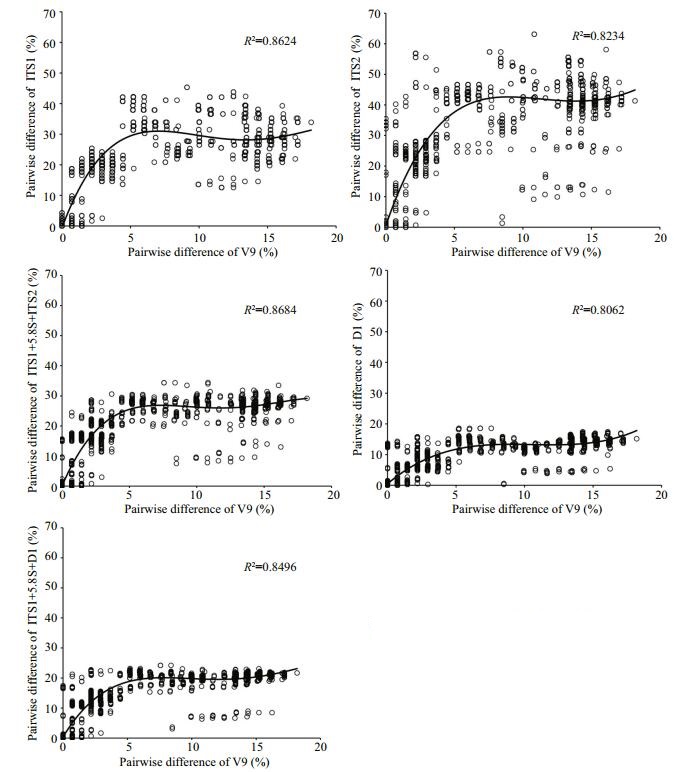
3.3 Relative variability of ITS and 28S regions versus 18S rDNA V9 regionIn our compiled datasets, the differences in the V9 regions ranged from 0 to 20% among the peritrichs (Fig. 3). The five rDNA loci (i.e., ITS1, ITS2, ITS, partial 28S and ITS+28S regions) co-varied well with the V9 regions, showing pairwise relationships that could be well predicted (R2 > 0.8; Fig. 3). With the increase of the difference in V9 regions from 0 to 5%, the corresponding variations of ITS1, ITS2, ITS, 28S and ITS+28S regions generally increased proportionally to 25%, 35%, 25%, 10% and 15%, respectively. Given the same variation of V9 region, the corresponding variations of ITS2 were much higher than other loci. It also showed the highest variation (35%) when the V9 variation was 5%. However, the differences of each of these five loci did not increase much when the variations of V9 were higher than 5%. As such, the variability of all the rDNA loci examined here appeared to be 2 to 7 folds higher than the V9 region, when the V9 region varies with a narrow range (< 5%).
|
| Figure 3 Scatter plots of pairwise differences between V9 and ITS1, ITS2, ITS1+ 5.8S+ITS2, D1 region of the 28S or ITS1+5.8S+ITS2+D1 Regression line and R2 were also shown. |
In order to obtained genetic information of peritrich ciliates from environmental samples, we have developed ITS-rDNA-targeted PCR primers in this study. We have showed evidence that the newly designed Peri1370F and PeriLsu1R are of high specificity in silico, for isolated specimens, and natural water and sediment samples. However, we do not claim that these primers are capable of covering all peritrich sequences at present. It is possible that the primer target most but not all peritrichs. For example, two strains of Zoothamnium and Epicarchesium are negatively amplified in our PCR tests for isolated specimens. Similar to the results of Liu and Gong (2012), sequences of several species like Epistylis galea, Campanella umbellaria, and Opercularia microdiscum were consistently lacking in the clone libraries of environmental samples. This might be due to the 1–3 mismatch of the forward primer Peri1370F to the 18S rDNA of the three taxa. Alternatively, these organisms might be absent in the samples tested. Pure isolates of these species can be tested to verify the specificity of the primer Peri1370F. Also, it is needed to bear in mind that we designed the reverse primer PeriLsu1R based on limited publically available information of 28S rDNA sequences from peritrichs. Currently, the amounts of peritrich 28S rRNA gene sequences are far fewer than those of the 18S in the GenBank, the validation and design of new primers of 28S need more reference sequences from identified peritrichs.
The results derived from the clone libraries provide some clues towards understanding the diversity and composition of peritrich ciliates in the environment. Firstly, the sequences of Vorticellidae were appeared to be abundant in the clone libraries of the water and sediment samples collected from the sea cucumber farming ponds, suggesting the taxon is probably abundant in terms of cell numbers. This is congruent with several morphology-based surveys reporting that Vorticella sp. was one of the dominant groups of the protozoa community in eutrophic habitats (Huang et al., 2005; Shi et al., 2012). Furthermore, some large sized peritrichs are known to have high rDNA copy numbers per cell (Gong et al., 2013; Fu and Gong, 2017), which may also account for the high proportions of vorticellid sequences in the clone libraries. Secondly, only few assemblages of sequences are shared between the sediment and water clone libraries (Fig. 2), suggesting different species composition in these two habitats. Interestingly, some peritrich species detected in our sediment libraries were found of ecological significance in various sediment samples. For example, Zoothamnium niveum and Pseudovorticella were described from the sediment of mangrove swamp. These species are known to live associate with bacterial sulphide-oxidizing ectosymbionts in sulphide rich mangrove swamp (Maurin et al., 2010). A fairly amount of our sediment sequences were affiliated to genus Pseudovorticella, which is known to be capable of cyst formation and was detected in the anoxic sediments dated to be at least 2 millions of years old (Orsi et al., 2013).
With the newly obtained V9, ITS and 28S rDNA data, our study shows that the full length of ITS, ITS1, ITS2 or D1 region of 28S are more variable than, and co-vary well with, the V9 region of 18S in some closely-related peritrichs (divergence < 5%). The V9 region has been used as a marker for metabarcoding of microbial eukaryotes including ciliates with high throughput sequencing technologies (Amaral-Zettler et al., 2009). This study illustrates the possibility to use more variable loci in resolving ecotypes or intraspecies distinctness within some widely distributed peritrich species. Among these, the ITS2 region seems to be a promising candidate for peritrichs owing to its high variability. This finding is congruent with a previous study, which suggests that the ITS2 is a suitable barcode for peritrich ciliates (Sun et al., 2012). Furthermore, our primer sets amplify an array of V9, ITS and partial 28S, which potentially link the existing data of V9 region to the more variable loci that offer additional information of intraspecies variations, thus provide a powerful tool for studying diversity and biogeography of peritrichs in diverse ecosystems.
Electronic supplementary material
Supplementary material (Supplementary Tables S1–S2) is available in the online version of this article at https://doi.org/10.1007/s00343-018-6326-3.
Altschul S F, Madden T L, Schäffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. 1997. Gapped BLAST and PSIBLAST:a new generation of protein database search programs. Nucleic Acids Research, 25(17): 3389-3402. DOI:10.1093/nar/25.17.3389 |
Amaral-Zettler L A, McCliment E A, Ducklow H W, Huse S M. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One, 4(7): e6372. DOI:10.1371/journal.pone.0006372 |
Bachy C, Dolan J R, López-García P, Deschamps P, Moreira D. 2013. Accuracy of protist diversity assessments:morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. The ISME Journal, 7(2): 244-255. DOI:10.1038/ismej.2012.106 |
Clamp J C, Coats D W. 2000. Planeticovorticella finleyi n.g., n.sp. (Peritrichia, Vorticellidae), a planktonic ciliate with a polymorphic life cycle. Invertebrate Biology, 119(1): 1-16. |
Foissner W, Agatha S, Berger H. 2002. Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib Desert. Biologiezentrum des Oberösterreichischen Landesmuseums, Linz, Austria. 1 459p.
|
Foissner W, Berger H, Kohmann F. 1991. Taxonomische und okologische Revision der Ciliaten des Saprobiensystems.Band Ⅱ:Peritrichia, Heterotrichida, Odontostomatida. Informationsberichte des Bayerischen Landesamtes für Wasserwirtschaft, 5/92: 1-502. |
Fried J, Mayr G, Berger H, Traunspurger W, Psenner R, Lemmer H. 2000. Monitoring protozoa and metazoa biofilm communities for assessing wastewater quality impact and reactor up-scaling effects. Water Science and Technology, 41(4-5): 309-316. DOI:10.2166/wst.2000.0460 |
Fu R, Gong J. 2017. Single cell analysis linking ribosomal (r)DNA and rRNA copy numbers to cell size and growth rate provides insights into molecular protistan ecology. Journal of Eukaryotic Microbiology. |
Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN:two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics, 12(6): 543-548. DOI:10.1093/bioinformatics/12.6.543 |
Gentekaki E, Lynn D. 2012. Spatial genetic variation, phylogeography and barcoding of the peritrichous ciliate Carchesium polypinum. European Journal of Protistology, 48(4): 305-313. DOI:10.1016/j.ejop.2012.04.001 |
Gong J, Dong J, Liu X H, Massana R. 2013. Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist, 164(3): 369-379. DOI:10.1016/j.protis.2012.11.006 |
Guo X H, Zhang Q Q, Zhang X L, Zhang J S, Gong J. 2015. Marine fungal communities in water and surface sediment of a sea cucumber farming system:habitat-differentiated distribution and nutrients driving succession. Fungal Ecology, 14: 87-98. DOI:10.1016/j.funeco.2014.12.001 |
Hall T A. 1999. BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41(2): 95-98. |
Hillis D M, Dixon M T. 1991. Ribosomal DNA:molecular evolution and phylogenetic inference. The Quarterly Review of Biology, 66(4): 411-453. DOI:10.1086/417338 |
Huang J R, Lin W H, Zeng W, Xu R L. 2005. The effect of sediment restoration on the protozoan community in shrimp culture ponds. Ecologic Science, 24(4): 326-329. |
Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon:a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics, 20(14): 2317-2319. DOI:10.1093/bioinformatics/bth226 |
Ji D D, Song W B, Al-Rasheid K A S, Li L F. 2005. Taxonomic characterization of two marine peritrichous ciliates, Pseudovorticella clampi n. sp. and Zoothamnium pararbuscula n. sp. (Ciliophora:Peritrichia), from North China. Journal of Eukaryotic Microbiology, 52(2): 159-169. DOI:10.1111/jeu.2005.52.issue-2 |
Ji D D, Sun P, Warren A, Song W B. 2009. Colonial sessilid peritrichs. In:Song W B, Warren A, Hu X eds. Free-living ciliates in the Bohai and Yellow Seas, China. Science Press, Beijing, China. p.257-286. (in Chinese)
|
Katoh K, Misawa K, Kuma K I, Miyata T. 2002. MAFFT:a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30(14): 3059-3066. DOI:10.1093/nar/gkf436 |
Liu X H, Gong J. 2012. Revealing the diversity and quantity of Peritrich ciliates in environmental samples using specific primer-based PCR and quantitative PCR. Microbes and Environments, 27(4): 497-503. DOI:10.1264/jsme2.ME12056 |
Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. 2008. probeCheck-a central resource for evaluating oligonucleotide probe coverage and specificity. Environmental Microbiology, 10(10): 2894-2898. DOI:10.1111/j.1462-2920.2008.01706.x |
Markmann M, Tautz D. 2005. Reverse taxonomy:an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Philosophical Transactions of the Royal Society B:Biological Sciences, 360(1462): 1917-1924. DOI:10.1098/rstb.2005.1723 |
Martín-Cereceda M, Serrano S, Guinea A. 2001. Biofilm communities and operational monitoring of a rotating biological contactor system. Water, Air, and Soil Pollution, 126(3-4): 193-206. |
Maurin L C, Himmel D, Mansot J L, Gros O. 2010. Raman microspectrometry as a powerful tool for a quick screening of thiotrophy:an application on mangrove swamp meiofauna of Guadeloupe (F.W.I.). Marine Environmental Research, 69(5): 382-389. DOI:10.1016/j.marenvres.2010.02.001 |
Orsi W, Biddle J F, Edgcomb V. 2013. Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One, 8(2): e56335. DOI:10.1371/journal.pone.0056335 |
Rocap G, Distel D L, Waterbury J B, Chisholm S W. 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Applied and Environmental Microbiology, 68(3): 1180-1191. DOI:10.1128/AEM.68.3.1180-1191.2002 |
Ronquist F, Huelsenbeck J P. 2003. MrBayes 3:bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1572-1574. DOI:10.1093/bioinformatics/btg180 |
Schloss P D, Westcott S L, Ryabin T, Hall J R, Hartmann M, Hollister E B, Lesniewski R A, Oakley B B, Parks D H, Robinson C J, Sahl J W, Stres B, Thallinger G G, Van Horn D J, Weber C F. 2009. Introducing mothur:opensource, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23): 7537-7541. DOI:10.1128/AEM.01541-09 |
Schoch C L, Seifert K A, Huhndorf S, Robert V, Spouge J L, Levesque C A, Chen W, Fungal Barcoding Consortium. 2012. Nuclear ribosomal internal transcribed spacer (ITS)region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America, 109(16): 6241-6246. DOI:10.1073/pnas.1117018109 |
Sherr B F, Sherr E B, Rassoulzadegan F. 1988. Rates of digestion of bacteria by marine phagotrophic protozoa:temperature dependence. Applied and Environmental Microbiology, 54(5): 1091-1095. |
Shi X L, Liu X J, Liu G J, Sun Z Q, Xu H L. 2012. An approach to analyzing spatial patterns of protozoan communities for assessing water quality in the Hangzhou section of Jing-Hang Grand Canal in China. Environmental Science and Pollution Research, 19(3): 739-747. DOI:10.1007/s11356-011-0615-0 |
Stamatakis A. 2014. RAxML version 8:a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9): 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Stern R F, Andersen R A, Jameson I, Küpper F C, Coffroth M A, Vaulot D, Le Gall F, Véron B, Brand J J, Skelton H, Kasai F, Lilly E L, Keeling P J. 2012. Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS One, 7(8). |
Sun P, Clamp J C, Xu D P, Huang B Q, Shin M K, Turner F. 2013. An ITS-based phylogenetic framework for the genus Vorticella:finding the molecular and morphological gaps in a taxonomically difficult group. Proceedings of the Royal Society B:Biological Science, 280(1771): 20131177. DOI:10.1098/rspb.2013.1177 |
Sun P, Clamp J, Xu D P, Kusuoka Y, Miao W. 2012. Vorticella Linnaeus, 1767 (Ciliophora, Oligohymenophora, Peritrichia) is a grade not a clade:redefinition of Vorticella and the families Vorticellidae and Astylozoidae using molecular characters derived from the gene coding for small subunit ribosomal RNA. Protist, 163(1): 129-142. DOI:10.1016/j.protis.2011.06.005 |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2725-2729. DOI:10.1093/molbev/mst197 |
Xu H L, Min G S, Choi J K, Jung J H, Park M H. 2009. An approach to analyses of periphytic ciliate colonization for monitoring water quality using a modified artificial substrate in Korean coastal waters. Marine Pollution Bulletin, 58(9): 1278-1285. DOI:10.1016/j.marpolbul.2009.05.003 |
| Altschul S F, Madden T L, Schäffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J, 1997. Gapped BLAST and PSIBLAST:a new generation of protein database search programs. Nucleic Acids Research, 25(17): 3389–3402. Doi: 10.1093/nar/25.17.3389 |
| Amaral-Zettler L A, McCliment E A, Ducklow H W, Huse S M, 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One, 4(7): e6372. Doi: 10.1371/journal.pone.0006372 |
| Bachy C, Dolan J R, López-García P, Deschamps P, Moreira D, 2013. Accuracy of protist diversity assessments:morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. The ISME Journal, 7(2): 244–255. Doi: 10.1038/ismej.2012.106 |
| Clamp J C, Coats D W, 2000. Planeticovorticella finleyi n.g., n.sp. (Peritrichia, Vorticellidae), a planktonic ciliate with a polymorphic life cycle. Invertebrate Biology, 119(1): 1–16. |
| Foissner W, Agatha S, Berger H. 2002. Soil ciliates (Protozoa, Ciliophora) from Namibia (Southwest Africa), with emphasis on two contrasting environments, the Etosha region and the Namib Desert. Biologiezentrum des Oberösterreichischen Landesmuseums, Linz, Austria. 1 459p. |
| Foissner W, Berger H, Kohmann F, 1991. Taxonomische und okologische Revision der Ciliaten des Saprobiensystems.Band Ⅱ:Peritrichia, Heterotrichida, Odontostomatida. Informationsberichte des Bayerischen Landesamtes für Wasserwirtschaft, 5/92: 1–502. |
| Fried J, Mayr G, Berger H, Traunspurger W, Psenner R, Lemmer H, 2000. Monitoring protozoa and metazoa biofilm communities for assessing wastewater quality impact and reactor up-scaling effects. Water Science and Technology, 41(4-5): 309–316. Doi: 10.2166/wst.2000.0460 |
| Fu R, Gong J, 2017. Single cell analysis linking ribosomal (r)DNA and rRNA copy numbers to cell size and growth rate provides insights into molecular protistan ecology. Journal of Eukaryotic Microbiology. |
| Galtier N, Gouy M, Gautier C, 1996. SEAVIEW and PHYLO_WIN:two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics, 12(6): 543–548. Doi: 10.1093/bioinformatics/12.6.543 |
| Gentekaki E, Lynn D, 2012. Spatial genetic variation, phylogeography and barcoding of the peritrichous ciliate Carchesium polypinum. European Journal of Protistology, 48(4): 305–313. Doi: 10.1016/j.ejop.2012.04.001 |
| Gong J, Dong J, Liu X H, Massana R, 2013. Extremely high copy numbers and polymorphisms of the rDNA operon estimated from single cell analysis of oligotrich and peritrich ciliates. Protist, 164(3): 369–379. Doi: 10.1016/j.protis.2012.11.006 |
| Guo X H, Zhang Q Q, Zhang X L, Zhang J S, Gong J, 2015. Marine fungal communities in water and surface sediment of a sea cucumber farming system:habitat-differentiated distribution and nutrients driving succession. Fungal Ecology, 14: 87–98. Doi: 10.1016/j.funeco.2014.12.001 |
| Hall T A, 1999. BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41(2): 95–98. |
| Hillis D M, Dixon M T, 1991. Ribosomal DNA:molecular evolution and phylogenetic inference. The Quarterly Review of Biology, 66(4): 411–453. Doi: 10.1086/417338 |
| Huang J R, Lin W H, Zeng W, Xu R L, 2005. The effect of sediment restoration on the protozoan community in shrimp culture ponds. Ecologic Science, 24(4): 326–329. |
| Huber T, Faulkner G, Hugenholtz P, 2004. Bellerophon:a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics, 20(14): 2317–2319. Doi: 10.1093/bioinformatics/bth226 |
| Ji D D, Song W B, Al-Rasheid K A S, Li L F, 2005. Taxonomic characterization of two marine peritrichous ciliates, Pseudovorticella clampi n. sp. and Zoothamnium pararbuscula n. sp. (Ciliophora:Peritrichia), from North China. Journal of Eukaryotic Microbiology, 52(2): 159–169. Doi: 10.1111/jeu.2005.52.issue-2 |
| Ji D D, Sun P, Warren A, Song W B. 2009. Colonial sessilid peritrichs. In:Song W B, Warren A, Hu X eds. Free-living ciliates in the Bohai and Yellow Seas, China. Science Press, Beijing, China. p.257-286. (in Chinese) |
| Katoh K, Misawa K, Kuma K I, Miyata T, 2002. MAFFT:a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30(14): 3059–3066. Doi: 10.1093/nar/gkf436 |
| Liu X H, Gong J, 2012. Revealing the diversity and quantity of Peritrich ciliates in environmental samples using specific primer-based PCR and quantitative PCR. Microbes and Environments, 27(4): 497–503. Doi: 10.1264/jsme2.ME12056 |
| Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M, 2008. probeCheck-a central resource for evaluating oligonucleotide probe coverage and specificity. Environmental Microbiology, 10(10): 2894–2898. Doi: 10.1111/j.1462-2920.2008.01706.x |
| Markmann M, Tautz D, 2005. Reverse taxonomy:an approach towards determining the diversity of meiobenthic organisms based on ribosomal RNA signature sequences. Philosophical Transactions of the Royal Society B:Biological Sciences, 360(1462): 1917–1924. Doi: 10.1098/rstb.2005.1723 |
| Martín-Cereceda M, Serrano S, Guinea A, 2001. Biofilm communities and operational monitoring of a rotating biological contactor system. Water, Air, and Soil Pollution, 126(3-4): 193–206. |
| Maurin L C, Himmel D, Mansot J L, Gros O, 2010. Raman microspectrometry as a powerful tool for a quick screening of thiotrophy:an application on mangrove swamp meiofauna of Guadeloupe (F.W.I.). Marine Environmental Research, 69(5): 382–389. Doi: 10.1016/j.marenvres.2010.02.001 |
| Orsi W, Biddle J F, Edgcomb V, 2013. Deep sequencing of subseafloor eukaryotic rRNA reveals active fungi across marine subsurface provinces. PLoS One, 8(2): e56335. Doi: 10.1371/journal.pone.0056335 |
| Rocap G, Distel D L, Waterbury J B, Chisholm S W, 2002. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Applied and Environmental Microbiology, 68(3): 1180–1191. Doi: 10.1128/AEM.68.3.1180-1191.2002 |
| Ronquist F, Huelsenbeck J P, 2003. MrBayes 3:bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1572–1574. Doi: 10.1093/bioinformatics/btg180 |
| Schloss P D, Westcott S L, Ryabin T, Hall J R, Hartmann M, Hollister E B, Lesniewski R A, Oakley B B, Parks D H, Robinson C J, Sahl J W, Stres B, Thallinger G G, Van Horn D J, Weber C F, 2009. Introducing mothur:opensource, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75(23): 7537–7541. Doi: 10.1128/AEM.01541-09 |
| Schoch C L, Seifert K A, Huhndorf S, Robert V, Spouge J L, Levesque C A, Chen W, Fungal Barcoding Consortium, 2012. Nuclear ribosomal internal transcribed spacer (ITS)region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America, 109(16): 6241–6246. Doi: 10.1073/pnas.1117018109 |
| Sherr B F, Sherr E B, Rassoulzadegan F, 1988. Rates of digestion of bacteria by marine phagotrophic protozoa:temperature dependence. Applied and Environmental Microbiology, 54(5): 1091–1095. |
| Shi X L, Liu X J, Liu G J, Sun Z Q, Xu H L, 2012. An approach to analyzing spatial patterns of protozoan communities for assessing water quality in the Hangzhou section of Jing-Hang Grand Canal in China. Environmental Science and Pollution Research, 19(3): 739–747. Doi: 10.1007/s11356-011-0615-0 |
| Stamatakis A, 2014. RAxML version 8:a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30(9): 1312–1313. Doi: 10.1093/bioinformatics/btu033 |
| Stern R F, Andersen R A, Jameson I, Küpper F C, Coffroth M A, Vaulot D, Le Gall F, Véron B, Brand J J, Skelton H, Kasai F, Lilly E L, Keeling P J, 2012. Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS One, 7(8). |
| Sun P, Clamp J C, Xu D P, Huang B Q, Shin M K, Turner F, 2013. An ITS-based phylogenetic framework for the genus Vorticella:finding the molecular and morphological gaps in a taxonomically difficult group. Proceedings of the Royal Society B:Biological Science, 280(1771): 20131177. Doi: 10.1098/rspb.2013.1177 |
| Sun P, Clamp J, Xu D P, Kusuoka Y, Miao W, 2012. Vorticella Linnaeus, 1767 (Ciliophora, Oligohymenophora, Peritrichia) is a grade not a clade:redefinition of Vorticella and the families Vorticellidae and Astylozoidae using molecular characters derived from the gene coding for small subunit ribosomal RNA. Protist, 163(1): 129–142. Doi: 10.1016/j.protis.2011.06.005 |
| Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2725–2729. Doi: 10.1093/molbev/mst197 |
| Xu H L, Min G S, Choi J K, Jung J H, Park M H, 2009. An approach to analyses of periphytic ciliate colonization for monitoring water quality using a modified artificial substrate in Korean coastal waters. Marine Pollution Bulletin, 58(9): 1278–1285. Doi: 10.1016/j.marpolbul.2009.05.003 |
 2018, Vol. 36
2018, Vol. 36


