Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LIAO Mengxiang(廖梦香), ZHENG Jiao(郑娇), WANG Zhiyong(王志勇), WANG Yilei(王艺磊), ZHANG Jing(张静), CAI Mingyi(蔡明夷)
- Molecular cytogenetic of the Amoy croaker, Argyrosomus amoyensis (Teleostei, Sciaenidae)
- Chinese Journal of Oceanology and Limnology, 36(3): 842-849
- http://dx.doi.org/10.1007/s00343-018-6272-0
Article History
- Received Oct. 29, 2016
- accepted in principle Jan. 7, 2017
- accepted for publication Mar. 29, 2017
Sciaenidae is one of the most species-rich families in the order Perciformes, with approximately 67 genera and 283 fish species (Nelson et al., 2016). Sciaenids are mainly distributed in the Indian, Pacific and Atlantic Oceans (Longhurst and Pauly, 1987; Sasaki, 1996), and are important fishery resources worldwide, as well as candidates for aquaculture. Although Sciaenidae is considered a monophyletic group, great diversity in body shape and mouth position relative to different feeding patterns and life histories has been observed in several species (Chao and Musick, 1977).
Despite its economic importance and great species diversity, few cytogenetic studies have been reported in this fish group, and most used conventional Giemsa staining (Arai, 2011). However, the available cytogenetic data suggest that the karyotypic macrostructure is remarkably conserved in the family (Accioly and Molina, 2008). Karyotype stability is a common trend in marine fishes, but some hypervariable chromosomal regions have been found, particularly those represented by ribosomal sites or positions adjacent to them (de Mello Affonso and Galetti, 2005). Thus, the chromosomal mapping of ribosomal sequences has received increasing attention as a way to identify and quantify chromosome dynamics (Calado et al., 2014).
The Amoy croaker (Argyrosomus amoyensis) (Bleeker, 1863; Perciformes, Sciaenidae), once named Nibea miichthioides, is a carnivorous sciaenid native to the Indo-West Pacific (Sasaki, 2001). It has become a popular mariculture species along the southeastern coast of China because of its delicious taste and strong disease resistance (Meng, 1996; Jian et al., 2013). The karyotype of the Amoy croaker has been described only once, by Wang et al. (2002) with Giemsa staining. Therefore, in the present study, the chromosomal distribution of the nucleolar organizer regions (NORs) was examined in the Amoy croaker using Giemsa, silver nitrate impregnation (AgNO3), denaturation and propidium iodide (DPI), and 4', 6-diamidino-2-phenylindole (DAPI) staining. Moreover, fluorescence in situ hybridization (FISH) with 18S (major) rDNAs, 5S (minor) rDNAs, and TTAGGGn sequences (a general telomere sequence for vertebrates) was performed, as well as a selfgenomic in situ hybridization (self-GISH) procedure. These results were compared with the available cytogenetic data of other Sciaenid species to explore whether the fine structure of the karyotype was as stable as the macrostructure in this family, and to screen out candidate cytogenetic markers for cytotaxonomic and phylogenetic studies.
2 MATERIAL AND METHOD 2.1 Specimen collection and chromosome preparationsFive male and five female Amoy croaker individuals used in the present study were obtained alive from the breeding base of Jimei University in Ningde, Fujian, China. Mitotic chromosome preparations were made from the head kidneys as described previously (Wang et al., 2002). Additionally, part of a fin from each fish was cut out and fixed in absolute ethyl alcohol to extract genomic DNA.
2.2 Chromosome staining and bandingThe chromosomes were examined by Giemsa, DPI, Ag-, and DAPI staining. In the Giemsa staining, the chromosomes were stained with 10% Giemsa in phosphate buffer (pH 6.8) for 25 min and then washed with running water. Ag-staining was performed as described by Howell and Black (1980). DPI staining was performed as described by Rab et al. (1996). For DAPI staining, the chromosomes were stained with 1 μg/mL DAPI in phosphate buffered saline (PBS, pH 7.0) for 10 min. C-banding was carried out according to Sumner's description (Sumner, 1972).
2.3 FISH and self-GISH proceduresGenomic DNA was extracted from the fixed fin tissue using a Genomic DNA Kit (Generay, Shanghai, China). A partial coding region of the 18S rDNA of A. amoyensis was amplified using the primers F (5'-CGCGCAAATTACCCACTCCC-3') and R (5'-CTGAACGCCACTTGTCCCT-3'), which were designed from a conserved region of the 18S rDNA from several fish species. The PCR reaction was performed in a total volume of 20 μL containing double distilled water, 1×PCR buffer, 0.2 mmol/L dNTPs, 0.5 mmol/L of each primer, 1 U of EasyTaq polymerase (TaKaRa, Shiga, Japan) and 50 ng genomic DNA. The PCR cycling conditions were an initial denaturation step at 94℃ for 4 min, followed by 30 cycles of 94℃ for 30 s, 54℃ for 1 min, 72℃ for 1 min, and a final extension step of 72℃ for 5 min. The amplified products were visualized after electrophoresis on a 1% agarose gel and sequenced for verification by a custom service (Shenggong, Shanghai, China). The whole coding and nontranscribed region of the 5S rDNA was obtained by PCR amplification with the primers F (5'-GTCAGGCCTGGTTAGTACTTGGAT-3') and R (5'- GGGCGCATTCAGGGTGGTAT-3'), which were designed from the coding sequence of the 5S rDNA from several fish species. PCR was performed as described above. The obtained PCR product was cloned into the pEASY-T1 vector (TransGen Biotech, Beijing, China) and sequenced for verification by a custom service (Shenggong). Telomeric repeats (TTAGGG)n were amplified by PCR without a template using (TTAGGG)5 and (TAACCC)5 primers (Ijdo et al., 1991). The extracted genomic DNA was used as probe in self-GISH experiments.
FISH and self-GISH were performed as described previously (Fujiwara et al., 1997; Cai et al., 2013). The 18S rDNA, 5S rDNA and genomic DNA probes were labeled with digoxigenin-11-dUTP or biotin-11- dUTP by nick translation according to the manufacturer's instructions (Roche, Basel, Switzerland). Chromosomes slides were denatured for 2 min in 70℃ 70% formamide/2× SSC, and then dehydrated in an ethanol series. Hybridization was performed with a mixture containing 2 ng/μL denatured probes without blocking DNAs, 50% deionized formamide, 10% dextran sulfate, 2×SSC, and double-deionized water at 37℃ for 8-16 h in a moist chamber. Post-hybridization washes were performed in 50% formamide/2×SSC at 37℃ for 20 min, 2×SSC and 1×SSC at room temperature for 20 min each, and 4×SSC at room temperature for 5 min. The digoxigenated and biotinylated probes were detected with anti-digoxigenin-rhodamine (Roche) and avidin-Alexa fluor-488 (Invitrogen, Carlsbad, CA, USA), respectively. Then, the chromosomes were counterstained with DAPI or PI in Antifade solution (Vector Laboratories, Burlingame, CA, USA).
2.4 Microscopy and image analysesGood chromosome samples at metaphase were examined and photographed using an Olympus BX53 fluorescence microscope (Olympus, Tokyo, Japan) and Olympus DP 80 digital image capture system (Olympus). Ten good metaphase plates without overlapping were analyzed. The relative length of each chromosome was measured with the Image-Pro Plus (IPP) 6.0 software, and the data were analyzed statistically as described previously (Cao et al., 2015). The chromosomes were classified according to Levan et al. (1964).
3 RESULTIn all 10 individuals examined, the Amoy croaker karyotype consisted of 48 acrocentric chromosomes. The chromosome complements were paired, and then ranked according to their average relative lengths (Fig. 1). The relative lengths were calculated based on data from 10 well-spread metaphases after self-GISH, which showed a continuous distribution. No obvious difference was found between the karyotypes of two sexes. After self-GISH, strong signals were present at the centromeric regions of all chromosomes (Fig. 1a).

|
| Figure 1 Karyotype of A. amoyensis a. self-GISH; b. C-banding. Bars represent 5 μm. |
C-banding revealed that the heterochromatic blocks not only located in the centromeric region, but also on NORs and in interstitial and telomeric regions of specific chromosomes. In addition, a greater variation in heterochromatin content was observed among chromosome pairs (Fig. 1b).
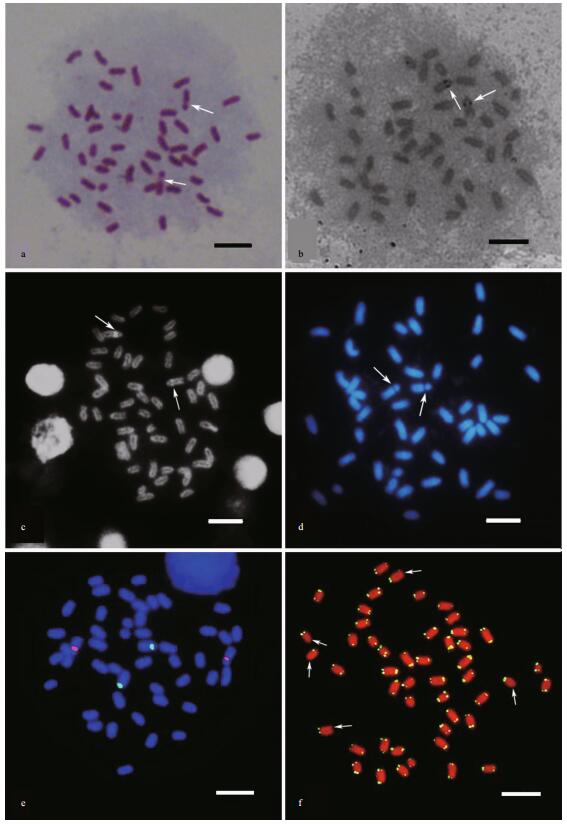
After Giemsa staining, the largest chromosome pair was easily distinguished by specific secondary constrictions at the proximal region, and was assigned to chromosome 1 (Fig. 2a). With AgNO3 staining, a single pair of active nucleolus organizer regions (AgNORs) was mapped to a position similar to the secondary constrictions (Fig. 2b). The NORs were also differentiated by DPI and DAPI staining (Fig. 2c, d), presenting as positive and negative bands (DPI+ and DAPI-), respectively. Size heteromorphism of the NORs between homologous chromosomes was observed with the above methods.
|
| Figure 2 Metaphase of A. amoyensis a. Giemsa staining, arrows indicate the secondary constriction; b. silver staining, arrows indicate AgNORs; c. DPI staining, arrows indicate positive areas; d. DAPI staining, arrows indicate negative bands; e. dual-color FISH with 18S rDNA (red) and 5S rDNA (green) probes; f. FISH with the telomeric sequence, arrows indicate chromosomes with a weak signal. Bars represent 5 μm. |
Dual-color FISH clearly showed that the 18S (red) and 5S (green) rDNA sites were located in different chromosome pairs (Fig. 2e). The 18S rDNA probes revealed two clusters corresponding to the Ag-NORs. The 5S rDNA sites were localized to the proximal region of chromosome 3. Size heteromorphism for both rDNA sites between homologous chromosomes was common in the observed metaphases. FISH with a telomere probe showed signals at both ends of all chromosomes, although some signals were very weak. Interstitial telomere signals were not found (Fig. 2f).
4 DISCUSSIONThe family Sciaenidae is remarkable for its chromosomal stability (Accioly and Molina, 2008). The representatives of this family share several symplesiomorphic cytogenetic features, such as 2n=48, fundamental number (FN)=48, a typical symmetrical karyotype, single and pericentromeric nucleolar organizer regions (NORs) and few conspicuous heterochromatic blocks (Accioly and Molina, 2008). This trend of chromosomal conservation, both numerical and structural, was corroborated by the present results obtained in the sciaenid A. amoyensis.
The karyotype of A. amoyensis comprised 2n=48 acrocentric chromosomes. This karyotype is a synapomorphy for most studied Sciaenids, expect a few species vary in chromosome numbers or fundamental numbers, such as Cynoscion arenarius (2n=48, FN=50), Corvina nigra (2n=46, FN=48) (Arai, 2011). A 48 acrocentric chromosome karyotype, also widespread in other families of Perciformes, is regarded as the primitive condition for this fish group (Galetti et al., 2000; Junior et al., 2006) and a synapomorphy for modern teleosteans (Brum and Galetti, 1997).
In A. amoyensis, the NORs were located at the proximal position of chromosome 1, which was confirmed by FISH with 18S rDNA probes. Interstitial NOR sites in a single acrocentric pair are also present in other sciaenid species (Table 1), and represent a conserved condition for this family, as well as for Perciformes (Aguilar and Galetti, 1997). Besides being interstitially located, NORs are also located in terminal regions in a few sciaenids, such as Larimichthys crocea and Ophioscion punctatissimus (Table 1). These variation of NOR location, together with the changes of FN in some sciaenids, indicate that pericentromeric rearrangements may have occurred occasionally with low frequency during the evolutionary process of sciaenids.
The 5S rDNA sites have been mapped in only four sciaenid species. Contrary to the stability of numbers of NORs, high levels of interspecific variation have been observed for 5S rDNA sites (Table 1). High dynamism of 5S rDNA sites is also present in other fish groups. For example, in the genus Gymnotus (Gymnotiformes, Gymnotidae), the 5S rDNA sites show different topologies and frequencies (from one pair in G. sylvius up to 15 in G. cf. carapo), although the 18S rDNA sites are conserved in both number and location (Scacchetti et al., 2011). The diversity of 5S rDNA together with relative stability of 18S rDNA in the karyotypes of sciaenids reinforces the hypothesis that they evolve independently and are subject to individual selective pressure (Martins and Galetti, 1999). Unequal exchange and transposition mediated by transposons were proposed as possible mechanisms for the high dynamism of rDNA (Coen and Dover, 1983; Da Silva et al., 2011; Symonová et al., 2013). Although the exact mechanism underlying the variability in the 5S rDNA sites of sciaenids is still unknown, a terminal distribution, as observed in A. amoyensis, S. ocellatus and L. crocea, may contribute to the dispersion of 5S rDNA sites. The dynamic character of 5S rDNA sites makes them effective cytotaxonomic markers, particularly under the background of low karyotypic diversity among sciaenids.
Dual-FISH showed that the 18S and 5S rDNA sites were located on distinct chromosome pairs in A. amoyensis. Divergent locations for the 18S and the 5S rDNA loci appear to be the most common situation in fish, and even in higher eukaryotes (Drouin and de Sá, 1995). The separate locations of the two rDNA clusters enable them to evolve independently, and avoid disruptive interference such as translocation between the 45S and 5S rDNA arrays. However, a syntenic organization for both rDNA sites has also been found in species in different groups, indicating that the conversion between synteny and divergence of 45S and 5S rDNA sites may have occurred independently many times during the evolutionary process of organisms (Martins and Galetti, 1999). For example, one of the two detected 5S rDNA sites were syntenic with the 45S rDNA in N. albiflora (Zheng et al., 2016).
Self-GISH is a modified GISH procedure in which self-genomic DNA probes free of blocking DNA are hybridized to the chromosomes. The procedure has been used to produce signal bands in plants as well as in fish (She et al., 2007; Markova and Vysko, 2009; Zheng et al., 2016). The signal pattern of self-GISH was demonstrated to be in accordance with that obtained from FISH with C0t-1 DNA in rice, suggesting that the signal intensity of self-GISH may reflect the C0t values of DNA probes (She et al., 2007). Hence, the self-GISH procedure has been used to survey the distribution of repetitive DNAs in chromosome complements and to assist chromosome identification (She et al., 2007; Zheng et al., 2016). In the present study, remarkably strong signals produced by self-GISH were distributed at all centromeric regions. However, the distribution of self-GISH signals is different from the C-bands, the regions of constitutive heterochromatin. Although the mechanism of GISH banding remains to be elucidated, the stable fluorescence bands obtained by the procedure can serve as useful markers for chromosome identification. It may be useful for sciaenids whose chromosomes are difficult to be distinguished for small size, similar shape and continuous distribution of quantitative characteristics (i.e. length).
The chromosomal distribution of the telomeric DNA sequences has been examined with FISH in approximately 80 fish species to investigate chromosomal rearrangements (Ocalewicz, 2013). A lack of interstitial telomere signals indicates chromosome rearrangement, inversion and fusion events may not have occurred during the evolutionary history of A. amoyensis, which reinforces de chromosomal conservation in this species (Slijepcevic, 1998; Ruiz-Herrera et al., 2008). The variability of signal intensity among the telomeres may reflect different telomere lengths among the chromosomes (Canela et al., 2007).
5 CONCLUSIONIn the present study, we updated the information on the karyotype of Amoy croaker. The karyotype of Amoy croaker was relatively conserved and primitive, considering its chromosome composition (2n=48a), single 18S rDNA at proximal region, no synteny between 18S and 5S rDNA sites, and no interstitial telomere. In comparison with the available cytogenetic data of other sciaenids, it can be deduced that although the karyotypic macrostructure and chromosomal location of 18S rDNA is predominantly conserved, sciaenids show some cytogenetic variability at the microstructural level. Therefore, critical analysis of the fine structures of the karyotypes with more markers in more sciaenid species is needed to investigate the dynamics of genes and repetitive sequences and to answer evolutionary and ecological questions.
Accioly I V, Molina W F. 2008. Cytogenetic studies in Brazilian marine Sciaenidae and Sparidae fishes (Perciformes). Genet. Mol. Res., 7(2): 358-370. DOI:10.4238/vol7-2gmr427 |
Aguilar C T, Galetti Jr P M. 1997. Chromosomal studies in South Atlantic serranids (Pisces, Perciformes). Cytobios, 89: 105-114. |
Arai R. 2011. Fish Karyotypes: A Check List. Springer, Tokyo, Japan. p. 174-175.
|
Brum M J I, Galetti Jr P M. 1997. Teleostei ground plan karyotype. J. Comp. Biol., 2: 91-102. |
Cai M Y, Liu X D, Chen Z Y, Cai B B, Ke C H. 2013. Characterization of Pacific abalone (Haliotis discus hannai) karyotype by C-banding and fluorescence in situ hybridization with rDNA. J. Fish. China, 37(7): 1002-1008. DOI:10.3724/SP.J.1231.2013.38481.(inChinesewithEnglishabstract) |
Calado L L, Bertollo L A C, Cioffi M B, Costa G W, Jacobina U P, Molina W F. 2014. Evolutionary dynamics of rDNA genes on chromosomes of the Eucinostomus fishes:cytotaxonomic and karyoevolutive implications. Genet.Mol. Res., 13(4): 9951-9959. DOI:10.4238/2014.November.27.24 |
Canela A, Vera E, Klatt P, Blasco M A. 2007. High-throughput telomere length quantification by fish and its application to human population studies. Proc. Natl. Acad. Sci. USA., 104(13): 5300-5305. DOI:10.1073/pnas.0609367104 |
Cao K, Zheng J, Wang Z Y, Liu X D, Cai M Y. 2015. Genome size and physical length of chromosomes in Nibea albiflora. South China Fish. Sci., 11(4): 65-70. DOI:10.3969/j.issn.2095-0780.2015.04.010.(inChinesewithEnglishabstract) |
Chao L N, Musick J A. 1977. Life history, feeding habits, and functional morphology of juvenile sciaenid fishes in the York River estuary, Virginia. Fish. Bull., 75(4): 657-702. |
Coen E S, Dover G A. 1983. Unequal exchanges and the coevolution of X and Y rDNA arrays in Drosophila melanogaster. Cell, 33(3): 849-855. DOI:10.1016/0092-8674(83)90027-2 |
Da Silva M, Matoso D A, Vicari M R, de Almeida M C, Margarido V P, Artoni R F. 2011. Physical mapping of 5S rDNA in two species of knifefishes:Gymnotus pantanal and Gymnotus paraguensis (gymnotiformes). Cytogenet.Genome Res., 134(4): 303-307. DOI:10.1159/000328998 |
de Mello Affonso P R A, Galetti Jr P M. 2005. Chromosomal diversification of reef fishes from genus Centropyge(perciformes, pomacanthidae). Genetica, 123(3): 227-233. DOI:10.1007/s10709-004-3214-x |
Drouin G, de Sá M M. 1995. The concerted evolution of 5S ribosomal genes linked to the repeat units of other Multigene Families. Mol. Biol. Evol., 12(3): 481-493. DOI:10.1093/oxfordjournals.molbev.a040223 |
Feldberg E, Porto J I R, dos Santos E B P, Valentim F C S. 1999. Cytogenetic studies of two freshwater sciaenids of the genus Plagioscion (Perciformes, sciaenidae) from the central Amazon. Genet. Mol. Biol., 22(3): 351-356. DOI:10.1590/S1415-47571999000300011 |
Fujiwara A, Abe S, Yamaha E, Yamazaki F, Yoshida M C. 1997. Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma, 106(1): 44-52. DOI:10.1007/s004120050223 |
Galetti Jr P M, Aguilar C T, Molina W F. 2000. An Overview of Marine Fish Cytogenetics. Springer, Netherlands. p. 55-62, https://doi.org/ 10.1007/978-94-017-2184-4_6.
|
Gomes V, Vazzoler A E A D M, Phan V N. 1983a. Estudos cariotípicos de peixes da família Sciaenidae (Teleostei, Perciformes) da região de Cananéia, SP, Brasil. 1. Sobre o cariótipo de Micropogonias furnieri (Desmarest, 1823). Bol. Inst. Oceanogr., 32(2): 137-142. DOI:10.1590/S0373-55241983000200004 |
Gomes V, Vazzoler A E A D M, Phan V N. 1983b. Estudos cariotípicos de peixes da família Sciaenidae (Teleostei, Perciformes) da região de Cananéia, SP, Brasil:2. Sobre o cariótipo de Menticirrhus americanus (Linnaeus, 1758). Bol. Inst. Oceanogr., 32(2): 187-191. DOI:10.1590/S0373-55241983000200009 |
Gornung E. 2013. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts:a review of research. Cytogenet. Genome Res., 141: 90-102. DOI:10.1159/000354832 |
Howell W M, Black D A. 1980. Controlled silver-staining of nucleolus organizer regions with protective colloidal developer:a 1-step method. Experientia, 36(8): 1014-1015. DOI:10.1007/BF01953855 |
Ijdo J W, Wells R A, Baldini A, Reeders S T. 1991. Improved telomere detection using a telomere repeat probe(TTAGGG)n generated by PCR. Nucleic Acids Res., 19(17): 4780. DOI:10.1093/nar/19.17.4780 |
Jian L J, Yang Y K, Liu X D, Chen Q K, Wang Z Y. 2013. The cross breeding and genetic analysis of hybrids of Larimichthys crocea (♀) and Nibea miichthioides (♂). J.Fish. China, 37(6): 801-808. DOI:10.3724/SP.J.1231.2013.38438.(inChinesewithEnglishabstract) |
Junior P M G, Molina W F, Affonso P R A M, Aguilar C T. 2006. Assessing genetic diversity of Brazilian reef fishes by chromosomal and DNA markers. Genetica, 126(1-2): 161-177. DOI:10.1007/s10709-005-1446-z |
Levan A, Fredga K, Sandberg A A. 1964. Nomenclature for centromeric position on chromosomes. Hereditas, 52(2): 201-220. DOI:10.1111/j.1601-5223.1964.tb01953.x |
Longhurst A R, Pauly D. 1987. Ecology of Tropical Oceans. Academic Press Inc., San Diego. p. 407.
|
Markova M, Vyskot B. 2009. New horizons of genomic in situ hybridization. Cytogenet. Genome Res., 126(4): 368-375. DOI:10.1159/000275796 |
Martins C, Galetti Jr P M. 1999. Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Res., 7(5): 363-367. DOI:10.1023/A:1009216030316 |
Meng Q W. 1996. Systematics of Fishes. China Agriculture Press, Beijing, China. p. 721-722. (in Chinese).
|
Nelson J S, Grande T C, Wilson M V H. 2016. Fishes of the World. 5th edn. John Wiley and Sons Inc., New York. p. 498.
|
Ocalewicz K. 2013. Telomeres in fishes. Cytogenet. Genome Res., 141(2-3): 114-125. DOI:10.1159/000354278 |
Ojima Y, Kikuno T. 1987. Karyotypes of a Gobiesociform and two Perciform fishes (Teleostei). Proc. Japan Acad. Ser.B, 63(6): 201-204. DOI:10.2183/pjab.63.201 |
Pereira A, Bedó G, Pereira J. 1988. Estudio cromosomico preliminar de Micropogonias furnieri Desmarest, 1823(Perciformes, Sciaenidae). Bol. Soc. Zool. Uruguay (2a epoca), 4: 23-26.
|
Rab P, Reed K M, de León A P, Phillips R B. 1996. A new method for detecting nucleolus organizer regions in fish chromosomes using denaturation and propidium iodide staining. Biotech. Histochem., 71(3): 157-162. DOI:10.3109/10520299609117153 |
Reggi R, Périco E, Suninsky M, Camillo J C A. 1986. Estudos citogenéticos em papa-terra, Menticirrhus litoralis(Perciformes, Serranidae). In: Simpósio de Citogenética Evolutiva e Aplicada de Peixes Neotropicais. Botucatu, UNESP. p. 57.
|
Ruiz-Herrera A, Nergadze S G, Santagostino M, Giulotto E. 2008. Telomeric repeats far from the ends:mechanisms of origin and role in evolution. Cytogenet. Genome Res., 122(3-4): 219-228. DOI:10.1159/000167807 |
Sasaki K. 1996. Sciaenid fishes of the Indian Ocean (Teleostei, Perciformes). Mem. Fac. Sci. Kochi. Univ. Ser. D, 16-17: 83-96. |
Sasaki K. 2001. Sciaenidae. Croakers (drums). In: Carpenter K E ed. FAO Species Identification Guide for Fishery Purposes. The Living Marine Resources of the Western Central Pacific. Vol. 5. Bony Fishes Part 3 (Menidae to Pomacentridae). Rome: FAO. p. 2 791-3 380.
|
Scacchetti P C, Pansonato-Alves J C, Utsunomia R, Oliveira C, Foresti F. 2011. Karyotypic diversity in four species of the genus Gymnotus Linnaeus, 1758 (Teleostei, Gymnotiformes, Gymnotidae):physical mapping of ribosomal genes and telomeric sequences. Comp Cytogen., 5(3): 223-235. DOI:10.3897/CompCytogen.v5i3.1375 |
She C W, Liu J Y, Diao Y, Hu Z L, Song Y C. 2007. The distribution of repetitive DNAs along chromosomes in plants revealed by self-genomic in situ hybridization. J.Genet. Genom., 34(5): 437-448. DOI:10.1016/S1673-8527(07)60048-4 |
Slijepcevic P. 1998. Telomeres and mechanisms of robertsonian fusion. Chromosoma, 107(2): 136-140. DOI:10.1007/s004120050289 |
Sumner A T. 1972. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res., 75(1): 304-306. DOI:10.1016/0014-4827(72)90558-7 |
Symonová R, Majtánová Z, Sember A, Staaks G B, Bohlen J, Freyhof J, Rábová M, Ráb P. 2013. Genome differentiation in a species pair of coregonine fishes:an extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol. Biol., 13(1): 42. DOI:10.1186/1471-2148-13-42 |
Wang D X, Wang J, Guo F, Liang J R, Qin Y X. 2002. Study on the karyotype in Nibea miichtheoides. Marine Sci, 26(11): 68-70. DOI:10.3969/j.issn.1000-3096.2002.11.019.(inChinesewithEnglishabstract) |
Wang X Y. 2012. Chromosome karyotypic analyses of some cultured fishes of the East China Sea. Zhejiang Ocean Univ. Nibea albiflora, Zhejiang, China. p. 1-65. (in Chinese).
|
Zheng J, Cao K, Yang A R, Zhang J, Wang Z Y, Cai M Y. 2016. Chromosome mapping using genomic DNA and repetitive DNA sequences as probes for somatic chromosome identification in Nibea albiflora. J. Fish. China, 40(8): 1156-1162. DOI:10.11964/jfc.20151110166.(inChinesewithEnglishabstract) |
| Accioly I V, Molina W F, 2008. Cytogenetic studies in Brazilian marine Sciaenidae and Sparidae fishes (Perciformes). Genet. Mol. Res., 7(2): 358–370. Doi: 10.4238/vol7-2gmr427 |
| Aguilar C T, Galetti Jr P M, 1997. Chromosomal studies in South Atlantic serranids (Pisces, Perciformes). Cytobios, 89: 105–114. |
| Arai R. 2011. Fish Karyotypes: A Check List. Springer, Tokyo, Japan. p. 174-175. |
| Brum M J I, Galetti Jr P M, 1997. Teleostei ground plan karyotype. J. Comp. Biol., 2: 91–102. |
| Cai M Y, Liu X D, Chen Z Y, Cai B B, Ke C H, 2013. Characterization of Pacific abalone (Haliotis discus hannai) karyotype by C-banding and fluorescence in situ hybridization with rDNA. J. Fish. China, 37(7): 1002–1008. Doi: 10.3724/SP.J.1231.2013.38481.(inChinesewithEnglishabstract) |
| Calado L L, Bertollo L A C, Cioffi M B, Costa G W, Jacobina U P, Molina W F, 2014. Evolutionary dynamics of rDNA genes on chromosomes of the Eucinostomus fishes:cytotaxonomic and karyoevolutive implications. Genet.Mol. Res., 13(4): 9951–9959. Doi: 10.4238/2014.November.27.24 |
| Canela A, Vera E, Klatt P, Blasco M A, 2007. High-throughput telomere length quantification by fish and its application to human population studies. Proc. Natl. Acad. Sci. USA., 104(13): 5300–5305. Doi: 10.1073/pnas.0609367104 |
| Cao K, Zheng J, Wang Z Y, Liu X D, Cai M Y, 2015. Genome size and physical length of chromosomes in Nibea albiflora. South China Fish. Sci., 11(4): 65–70. Doi: 10.3969/j.issn.2095-0780.2015.04.010.(inChinesewithEnglishabstract) |
| Chao L N, Musick J A, 1977. Life history, feeding habits, and functional morphology of juvenile sciaenid fishes in the York River estuary, Virginia. Fish. Bull., 75(4): 657–702. |
| Coen E S, Dover G A, 1983. Unequal exchanges and the coevolution of X and Y rDNA arrays in Drosophila melanogaster. Cell, 33(3): 849–855. Doi: 10.1016/0092-8674(83)90027-2 |
| Da Silva M, Matoso D A, Vicari M R, de Almeida M C, Margarido V P, Artoni R F, 2011. Physical mapping of 5S rDNA in two species of knifefishes:Gymnotus pantanal and Gymnotus paraguensis (gymnotiformes). Cytogenet.Genome Res., 134(4): 303–307. Doi: 10.1159/000328998 |
| de Mello Affonso P R A, Galetti Jr P M, 2005. Chromosomal diversification of reef fishes from genus Centropyge(perciformes, pomacanthidae). Genetica, 123(3): 227–233. Doi: 10.1007/s10709-004-3214-x |
| Drouin G, de Sá M M, 1995. The concerted evolution of 5S ribosomal genes linked to the repeat units of other Multigene Families. Mol. Biol. Evol., 12(3): 481–493. Doi: 10.1093/oxfordjournals.molbev.a040223 |
| Feldberg E, Porto J I R, dos Santos E B P, Valentim F C S, 1999. Cytogenetic studies of two freshwater sciaenids of the genus Plagioscion (Perciformes, sciaenidae) from the central Amazon. Genet. Mol. Biol., 22(3): 351–356. Doi: 10.1590/S1415-47571999000300011 |
| Fujiwara A, Abe S, Yamaha E, Yamazaki F, Yoshida M C, 1997. Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma, 106(1): 44–52. Doi: 10.1007/s004120050223 |
| Galetti Jr P M, Aguilar C T, Molina W F. 2000. An Overview of Marine Fish Cytogenetics. Springer, Netherlands. p. 55-62, https://doi.org/ 10.1007/978-94-017-2184-4_6. |
| Gomes V, Vazzoler A E A D M, Phan V N, 1983a. Estudos cariotípicos de peixes da família Sciaenidae (Teleostei, Perciformes) da região de Cananéia, SP, Brasil. 1. Sobre o cariótipo de Micropogonias furnieri (Desmarest, 1823). Bol. Inst. Oceanogr., 32(2): 137–142. Doi: 10.1590/S0373-55241983000200004 |
| Gomes V, Vazzoler A E A D M, Phan V N, 1983b. Estudos cariotípicos de peixes da família Sciaenidae (Teleostei, Perciformes) da região de Cananéia, SP, Brasil:2. Sobre o cariótipo de Menticirrhus americanus (Linnaeus, 1758). Bol. Inst. Oceanogr., 32(2): 187–191. Doi: 10.1590/S0373-55241983000200009 |
| Gornung E, 2013. Twenty years of physical mapping of major ribosomal RNA genes across the teleosts:a review of research. Cytogenet. Genome Res., 141: 90–102. Doi: 10.1159/000354832 |
| Howell W M, Black D A, 1980. Controlled silver-staining of nucleolus organizer regions with protective colloidal developer:a 1-step method. Experientia, 36(8): 1014–1015. Doi: 10.1007/BF01953855 |
| Ijdo J W, Wells R A, Baldini A, Reeders S T, 1991. Improved telomere detection using a telomere repeat probe(TTAGGG)n generated by PCR. Nucleic Acids Res., 19(17): 4780. Doi: 10.1093/nar/19.17.4780 |
| Jian L J, Yang Y K, Liu X D, Chen Q K, Wang Z Y, 2013. The cross breeding and genetic analysis of hybrids of Larimichthys crocea (♀) and Nibea miichthioides (♂). J.Fish. China, 37(6): 801–808. Doi: 10.3724/SP.J.1231.2013.38438.(inChinesewithEnglishabstract) |
| Junior P M G, Molina W F, Affonso P R A M, Aguilar C T, 2006. Assessing genetic diversity of Brazilian reef fishes by chromosomal and DNA markers. Genetica, 126(1-2): 161–177. Doi: 10.1007/s10709-005-1446-z |
| Levan A, Fredga K, Sandberg A A, 1964. Nomenclature for centromeric position on chromosomes. Hereditas, 52(2): 201–220. Doi: 10.1111/j.1601-5223.1964.tb01953.x |
| Longhurst A R, Pauly D. 1987. Ecology of Tropical Oceans. Academic Press Inc., San Diego. p. 407. |
| Markova M, Vyskot B, 2009. New horizons of genomic in situ hybridization. Cytogenet. Genome Res., 126(4): 368–375. Doi: 10.1159/000275796 |
| Martins C, Galetti Jr P M, 1999. Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Res., 7(5): 363–367. Doi: 10.1023/A:1009216030316 |
| Meng Q W. 1996. Systematics of Fishes. China Agriculture Press, Beijing, China. p. 721-722. (in Chinese). |
| Nelson J S, Grande T C, Wilson M V H. 2016. Fishes of the World. 5th edn. John Wiley and Sons Inc., New York. p. 498. |
| Ocalewicz K, 2013. Telomeres in fishes. Cytogenet. Genome Res., 141(2-3): 114–125. Doi: 10.1159/000354278 |
| Ojima Y, Kikuno T, 1987. Karyotypes of a Gobiesociform and two Perciform fishes (Teleostei). Proc. Japan Acad. Ser.B, 63(6): 201–204. Doi: 10.2183/pjab.63.201 |
| Pereira A, Bedó G, Pereira J. 1988. Estudio cromosomico preliminar de Micropogonias furnieri Desmarest, 1823(Perciformes, Sciaenidae). Bol. Soc. Zool. Uruguay (2a epoca), 4: 23-26. |
| Rab P, Reed K M, de León A P, Phillips R B, 1996. A new method for detecting nucleolus organizer regions in fish chromosomes using denaturation and propidium iodide staining. Biotech. Histochem., 71(3): 157–162. Doi: 10.3109/10520299609117153 |
| Reggi R, Périco E, Suninsky M, Camillo J C A. 1986. Estudos citogenéticos em papa-terra, Menticirrhus litoralis(Perciformes, Serranidae). In: Simpósio de Citogenética Evolutiva e Aplicada de Peixes Neotropicais. Botucatu, UNESP. p. 57. |
| Ruiz-Herrera A, Nergadze S G, Santagostino M, Giulotto E, 2008. Telomeric repeats far from the ends:mechanisms of origin and role in evolution. Cytogenet. Genome Res., 122(3-4): 219–228. Doi: 10.1159/000167807 |
| Sasaki K, 1996. Sciaenid fishes of the Indian Ocean (Teleostei, Perciformes). Mem. Fac. Sci. Kochi. Univ. Ser. D, 16-17: 83–96. |
| Sasaki K. 2001. Sciaenidae. Croakers (drums). In: Carpenter K E ed. FAO Species Identification Guide for Fishery Purposes. The Living Marine Resources of the Western Central Pacific. Vol. 5. Bony Fishes Part 3 (Menidae to Pomacentridae). Rome: FAO. p. 2 791-3 380. |
| Scacchetti P C, Pansonato-Alves J C, Utsunomia R, Oliveira C, Foresti F, 2011. Karyotypic diversity in four species of the genus Gymnotus Linnaeus, 1758 (Teleostei, Gymnotiformes, Gymnotidae):physical mapping of ribosomal genes and telomeric sequences. Comp Cytogen., 5(3): 223–235. Doi: 10.3897/CompCytogen.v5i3.1375 |
| She C W, Liu J Y, Diao Y, Hu Z L, Song Y C, 2007. The distribution of repetitive DNAs along chromosomes in plants revealed by self-genomic in situ hybridization. J.Genet. Genom., 34(5): 437–448. Doi: 10.1016/S1673-8527(07)60048-4 |
| Slijepcevic P, 1998. Telomeres and mechanisms of robertsonian fusion. Chromosoma, 107(2): 136–140. Doi: 10.1007/s004120050289 |
| Sumner A T, 1972. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res., 75(1): 304–306. Doi: 10.1016/0014-4827(72)90558-7 |
| Symonová R, Majtánová Z, Sember A, Staaks G B, Bohlen J, Freyhof J, Rábová M, Ráb P, 2013. Genome differentiation in a species pair of coregonine fishes:an extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol. Biol., 13(1): 42. Doi: 10.1186/1471-2148-13-42 |
| Wang D X, Wang J, Guo F, Liang J R, Qin Y X, 2002. Study on the karyotype in Nibea miichtheoides. Marine Sci, 26(11): 68–70. Doi: 10.3969/j.issn.1000-3096.2002.11.019.(inChinesewithEnglishabstract) |
| Wang X Y. 2012. Chromosome karyotypic analyses of some cultured fishes of the East China Sea. Zhejiang Ocean Univ. Nibea albiflora, Zhejiang, China. p. 1-65. (in Chinese). |
| Zheng J, Cao K, Yang A R, Zhang J, Wang Z Y, Cai M Y, 2016. Chromosome mapping using genomic DNA and repetitive DNA sequences as probes for somatic chromosome identification in Nibea albiflora. J. Fish. China, 40(8): 1156–1162. Doi: 10.11964/jfc.20151110166.(inChinesewithEnglishabstract) |
 2018, Vol. 36
2018, Vol. 36