Institute of Oceanology, Chinese Academy of Sciences
Article Information
- YANG Mei(杨梅), LI Xinzheng(李新正)
- Population genetic structure of the mantis shrimp Oratosquilla oratoria (Crustacea: Squillidae) in the Yellow Sea and East China Sea
- Chinese Journal of Oceanology and Limnology, 36(3): 905-912
- http://dx.doi.org/10.1007/s00343-018-7030-z
Article History
- Received Feb. 7, 2017
- accepted in principle Apr. 7, 2017
- accepted for publication May. 30, 2017
2 University of Chinese Academy of Sciences, Beijing 100049, China;
3 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266000, China
In Northwestern Pacific, the marginal seas of China has well-characterized oceanography and dramatic geological history, which provides one of the best model system for marine organisms population genetics research (Kong and Li, 2009). Recent studies revealed that the historical separation of the sea basins during the Pleistocene glaciers, coastal topography, ocean currents and freshwater outflow played discrete but important influences in shaping the present genetic structure of various marine species (Liu et al., 2007; Shen et al., 2011; Dong et al., 2012; Hurwood et al., 2014).
Oratosquilla oratoria is widely distributed in the Western Pacific, and has been regarded as one of the valuable fishery resources for its good taste and rich nutrition (Wang et al., 1996). In recent years, the natural resources of O. oratoria have been decreased rapidly due to overfishing, marine pollution and habitat destruction. Inevitably, the decline in wild resource will reduce the intraspecific genetic diversity and weakening the understanding of population genetics to the species (Lu et al., 2013). With stable maternal inheritance and a generally high rate of base substitutions, mitochondrial DNA (mtDNA) markers have been widely used for studies of intraspecific population genetics (de Bruyn et al., 2005; Alam et al., 2015). For example, mtDNA variation in Penaeus semisulcatus reveals two distinct lineages in the IndoWest Pacific region (Alam et al., 2017). And due to high commercial and ecological values of the O. oratoria, there were several researches to estimate the genetic diversity and genetic structure in China Sea (Lui et al., 2010; Zhang et al., 2012, 2016; Du et al., 2016). In present study, based on the analysis of mtDNA control region sequences, we surveyed the population genetics of O. oratoria in two marginal seas (i.e., the Yellow Sea and East China Sea) by intensive sampling. And we aimed to gain complementary information about the present situation of genetic diversity as well as evolutionary patterns of O. oratoria.
2 MATERIAL AND METHOD 2.1 Sample collectionAll the specimens of wild O. oratoria were collected between May 2014 and November 2015. We obtained 394 individuals of this species from 18 locations in the Yellow Sea and East China Sea, with a range of 12–35 individuals per site (Fig. 1 and Table 1). The materials were sampled and preserved in 95% ethanol for subsequent DNA extraction.

|
| Figure 1 Sampling locations of Oratosquilla oratoria Yellow Sea: Y1–Y4; East China Sea: E1–E14. The red line indicates the path of the Kuroshio Current and its tributaries in the East China Sea, N-KBBC: an nearshore Kuroshio bottom branch current; O-KBBC: an offshore Kuroshio bottom branch current. |
|
Genomic DNA was extracted from muscle tissue by the Tissue DNA Kit (OMEGA). For each individual, fragments of mtDNA control region were amplified with primer pairs: CR-F1 (5′-TAACCGCGACTGCTGGCAC-3′), CR-F2 (5′-GGGTATGAGCCCATTAGCTT-3′) (Lui et al., 2010). PCR amplifications were carried out in 25 μL volume containing 12.5 μL 2X Premix Ex Taq (TaKaRa Co., China), 1 μL of each primer, 1 μL template DNA and 9.5 μL ddH2O. The cycling conditions were as follows: initial denaturation at 94℃ for 2 min, followed by 30 cycles of 94℃ for 1 min, 56℃ for 45 s, 72℃ for 50 s, then a final extension at 72℃ for 10 min. PCR products were purified using the PCR clean-up Kit (Axygen) and sequenced on ABI 3730 automatic sequencer (Sangon Inc., China).
2.3 Data analysesSequences were aligned with ClustalX (Thompson et al., 1997) using default settings and corrected by hand. The nucleotide diversity (π) and haplotype diversity (h) were calculated using the DnaSP (Librado and Rozas, 2009). Phylogenies of haplotypes were reconstructed by the neighbor-joining (NJ) (Saitou and Nei, 1987) (Bootstrap=1 000) in MEGA 6.0. In addition, a median-joining (MJ) was generated for haplotypes using the Network (Bandelt et al., 1999). Population genetic structure was evaluated with FST statistics and AMOVA (analysis of molecular variance) (Excoffier et al., 1992) in ARLEQUIN (Excoffier and Lischer, 2010). The Tajima's D (Tajima, 1989) statistics, Fu's FS (Fu, 1997) test of neutrality and the frequency distribution of pairwise differences between mtDNA haplotypes (i.e., mismatch distribution) were conducted to infer the population demographic history of O. oratoria. The tau value (τ), which reflects the location of the mismatch distribution crest, provides a rough estimate of the time when rapid population range expansion started. According to t=τ/2u, calculate the expansion time of O. oratoria in the Yellow Sea and East China Sea, respectively. And the mutation rate of 3.44%/ Myr for Mitochondrial control region was applied to in the present study (Alam, 2016).
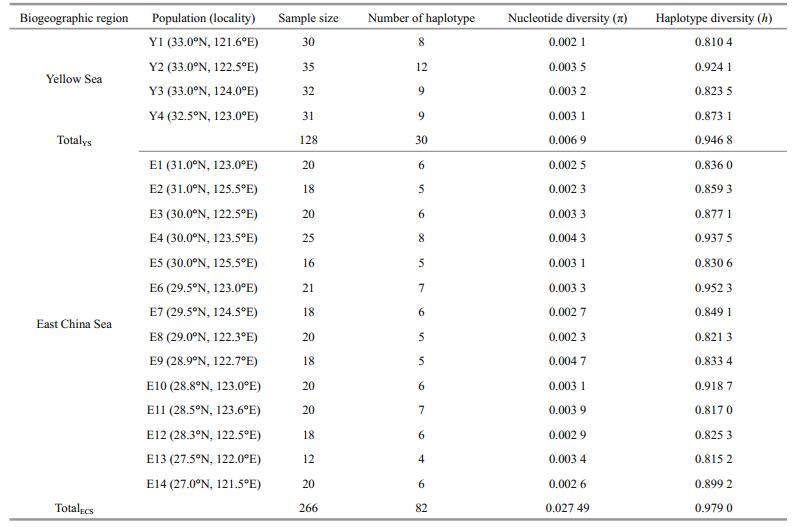
3 RESULT 3.1 Genetic diversityA 923-bp segment of mtDNA control region was amplified and total of 102 different haplotypes were identified based on the sequence variation in 394 individuals from 18 locations of the Yellow Sea and East China Sea. There were 30 haplotypes of 128 individuals detected in the Yellow Sea, while 82 haplotypes of 266 individuals in the East China Sea (Table 1). And among these, 10 of the haplotypes were shared. The most common haplotype, Hap6, was shared by a total of 62 individuals from the Yellow Sea and East China Sea.
The estimates of nucleotide diversity (π) and haplotype diversity (h) for different locations were showed in Table 1. The highest value of π was in E9 (0.004 7) of the East China Sea and the lowest was in Y1 (0.002 1) of the Yellow Sea; the highest value of h was in E6 (0.952 3) of the East China Sea and the lowest was in Y1 (0.810 4) of the Yellow Sea. For the Yellow Sea, the overall nucleotide diversity and haplotype diversity were 0.006 9 and 0.946 8, respectively; while across all the East China Sea locations, the overall nucleotide diversity and haplotype diversity were 0.027 94 and 0.979 0, respectively.
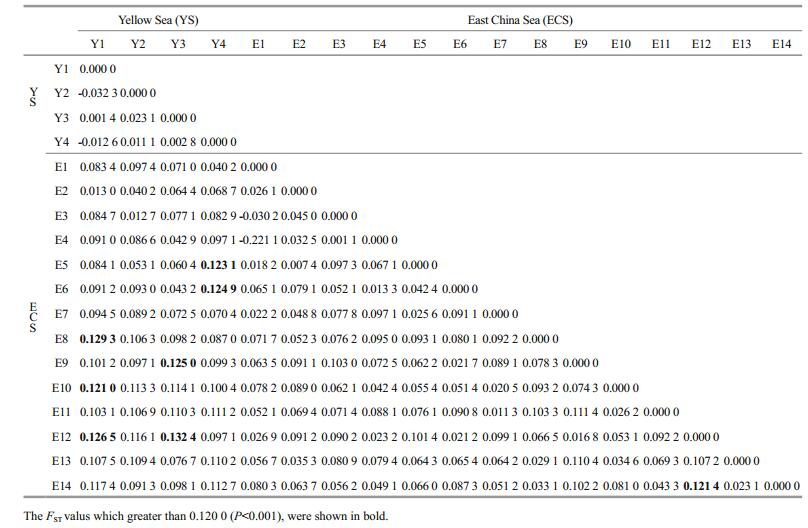
3.2 Population structureGenetic differentiation among eighteen different locations was evaluated using FST values and AMOVA analyses. In general, the pairwise FST were range from -0.032 3 to 0.132 4 (Table 2), the largest genetic differentiation occurred between Y3 and E12 (FST=0.132 4, P < 0.001). The pairwise FST values between the Yellow Sea (Y1–Y4) and East China Sea (E1–E14) populations was 0.145 2 (P < 0.001), indicated a moderate degree of differentiation was existed. Furthermore, we did an analysis of genetic differentiation between the Kuroshio tributary and other populations in the East China Sea. According to previous studies about Kuroshio in the East China Sea (Yang, 2011), we considered the individuals from E4, E6, E9, E10 and E12 as a population in Kuroshio tributary (N-KBBC: a nearshore Kuroshio bottom branch current), while remaining individuals (E1–E3, E5, E7–E8, E11, E13–E14) as the other population (Fig. 1). The result showed that differentiation existed between above populations (FST=0.0495, P < 0.001).
One-group AMOVA analysis showed that the overall population differentiation was 14.46% and genetic variation within populations was 85.54% (P < 0.001). Two groups AMOVA (Yellow Sea and East China Sea) showed that the source of genetic differentiation was mainly found within populations (89.5%), while the rest was distributed among populations within groups (7.2%) and among groups (3.3%).
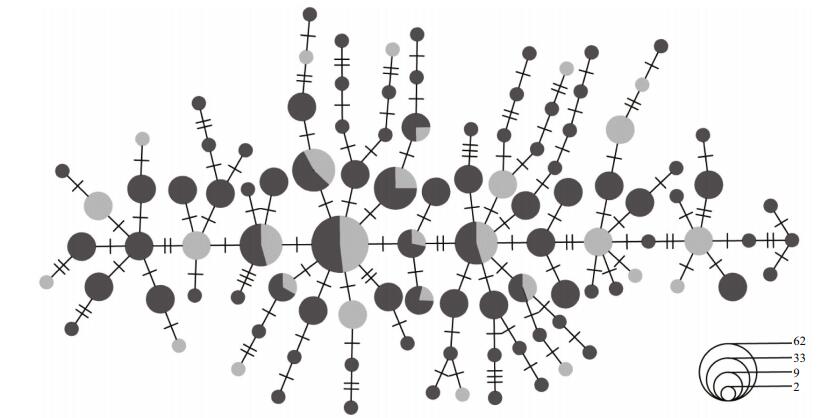
The median-joining network of O. oratoria proved that the haplotypes could not be clearly divided into different geographical groups, and was characterized by a star-like phylogeny with closely related haplotypes derived from the most common and ancestral haplotype (Fig. 2). Moreover, the topology of the NJ tree was shallow and displayed no significant genealogical branches of samples corresponding to sampling locations (data not presented).
|
| Figure 2 Median-joining network of haplotypes of Oratosquilla oratoria Sizes of circles are proportional to haplotype frequency. Perpendicular tick marks on the lines joining haplotypes represent the number of nucleotide substitutions. Yellow Sea (grey), East China Sea (black). |
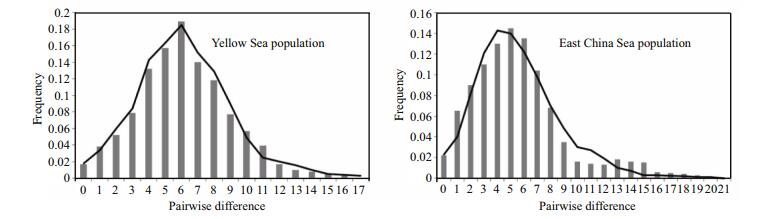
The mismatch distribution of O. oratoria for the Yellow Sea and East China Sea were consisted of a distinct unimodal curve, which closely fitted to the expected distributions under a rapid population expansion model (Fig. 3). In addition, both Tajima's D statistics (Tajima's DYS=-1.94, P < 0.001; Tajima's DECS=-2.29, P < 0.001) and Fu's FS test (FS–YS=-6.05; FS–ECS=-11.79) were all showed significant negative values. All of these results suggested that the O. oratoria had undergone a recent colonization and sudden population expansion events in the Yellow Sea and East China Sea. And the estimate of time of expansion for the Yellow Sea was 176 800 years, while for the East China Sea, this estimate was 206 600 years ago.
|
| Figure 3 Observed pairwise differences (bars), and the expected mismatch distributions under the sudden expansion model (line) for the control region in Oratosquilla oratoria |
Generally, due to high dispersal potential (planktonic egg, larval or adult history stages) and absence of physical barriers, marine organisms are expected to have low degree of genetic differentiation among geographical regions (Palumbi, 1994; Hewitt, 2000). However, in present study, the results of pairwise FST and AMOVA analyses revealed moderate population structure of O. oratoria between the Yellow Sea and East China Sea populations. But neither the neighbor-joining tree nor haplotype network showed clades with geographic pattern, which indicated effective gene flow was existed between these marginal seas. Considering the O. oratoria has a long planktonic larval phase of between 36 to 59 days (Hamano and Matsuura, 1987), larval dispersal driven by oceanographic circulation might play important role in maintenance of population connectivity. In the Yellow Sea and East China Sea, the composition of ocean currents circulation is complex, including China Coastal Current, Changjiang (Yangtze) River outflow, Kuroshio tributary and so on (Li et al., 2000). Previous studies have been confirmed that the China Coastal Current transport certain floating marine organisms from the Yellow Sea to the East/South China Sea, such as fragments of Sargassum and Calanus sinicus (Hwang and Wong, 2005; Cheang et al., 2010). Our results suggested that planktonic larvae of O. oratoria in the Yellow Sea and East China Sea might travel certain distances with ocean currents, yielding gene flow among different geographical regions.
Besides ocean currents, the dispersal of larval can also be affected by freshwater runoff from the Changjiang River. As a euryhaline species, O. oratoria appears to tolerance wide raged salinity (20– 40) but sensitive to low salinity (lower than 15) (Liu et al., 2006, 2012). Meanwhile, the peak spawing period (May to July) of O. oratoria was consist with the large discharge of Changjiang River in summer, which greatly reduced the seawater salinity in the estuary area (Wang et al., 1998). It is likely that, to some extent, the Changjiang River form a physical barrier, which acted to prevent transport of larval or adults of O. oratoria among different biogeographic regions. And the barrier effect of the Changjiang River outflow have been ascertained by relevant studies (Du et al., 2016). Additionally, in recent years there were several studies have focused on population genetic structure of marine species between the Yellow Sea and East China Sea (Ni et al., 2014). Genetic studies on Nibea albiflora, Portunus trituberculatus, Cyclina sinensis and Charybdis bimaculata found no significant genetic structure between the Yellow Sea and East China Sea populations, showed the Changjiang River outflow was not validated for these species because their prejuveniles and juveniles are able to survive over a wide range of salinities (Feng et al., 2008; Han et al., 2008, 2015a; Ni et al., 2012). However, similar researches on limpet Cellana toreuma and cocktail shrimp Trachypenaeus curvirostris indicated that significant genetic differentiation in line with the outflow, which limited the dispersal of larvae along the Chinese coast (Dong et al., 2012; Han et al., 2015b). These results revealed that the Changjiang River outflow affect some species depending on habitat specificity and differentiation in biological characteristics (such as salinity tolerance and dispersal ability) (Ni et al., 2014).
Furthermore, in our study, differentiation was detected between the Kuroshio tributary and other regions populations of the East China Sea. The Kuroshio Current originates in the westward-flowing North Equatorial Current of the central Pacific then deflects towards the northeast offshore of the Philippine Islands (Dou et al., 2012), and producing striking influence on biodiversity and genetic structure of marine species in the Northwestern Pacific (Yasuda et al., 2009; Nakajima et al., 2014). Along the coastline of Taiwan, the Kuroshio Current flows south to north at an average rate of 25 cm/s (Ju et al., 2013) and can carry larval of O. oratoria from the South China Sea to the East China Sea. Previous studies have showed the formation of Taiwan limited the gene flow of O. oratoria between the East China Sea and South China Sea (Zhang et al., 2014, 2016). Additionally, ocean currents of the East China Sea exhibited characteristics of counterclock wise cyclonic circulation, while the surface circulation of the South China Sea has clockwise circulation due to the monsoon drift, the opposite ocean currents driving the planktonic larvae of O. oratoria in opposite directions during the reproductive season further deepened their divergence (Zhang et al., 2014, 2016).
Evidence for the occurrence of population expansion of O. oratoria in the Yellow Sea and East China Sea were confirmed by the mismatch distribution analyses and neutrality tests (Fig. 3). Based on values of the nucleotide and haplotype diversity of mtDNA, four basic scenarios of population history were proposed by Grant and Bowen (1998). Oratosquilla oratoria with low nucleotide diversity and high haplotype diversity probably undergone sudden population expansion after a period of low effective population size. Once population size increases rapidly following environmental disturbances, haplotype diversity can usually recover via mutation, whereas nucleotide diversity remains low since there was not enough time to accumulate large sequence differences between haplotypes (Avise, 2000; Lui et al., 2010). According to previous phylogeographic studies, the population sudden expansion of marine organisms was common in the Northwestern Pacific (Ni et al., 2014). During the Pleistocene glacial cycles, as one of the most extensive continental shelves, the Yellow Sea and most of the East China Sea were exposed, and the latter was consequently reduced into an elongated enclosed sea with an area < 1/3 of its present size (Wang, 1999; Kitamura et al., 2001). The O. oratoria in these marginal seas can survived only in common glacial refuge regions, and in postglacial, when sea level rose as glaciers melted, the Yellow Sea and East China Sea must have been recolonized by O. oratoria. Furthermore, our studies revealed a shallow and star-like topology, which suggested an origin from a common ancestral population in the East China Sea refugium.
5 CONCLUSIONIn the present study, O. oratoria showed moderate degree of genetic differentiation between the Yellow Sea and East China Sea populations. The Changjiang River acted as a physical barrier to prevent transport of larval or adults of O. oratoria between these marginal seas, while the considerable gene flow was maintained due to the larval dispersal with ocean currents. And based on our results, the O. oratoria needs cautious to deal different stocks in the Yellow Sea and East China Sea. Further studies of O. oratoria throughout the whole Western Pacific and employ more molecular markers would contribution to enhancing the knowledge of the genetic structure and evolutionary history, thus provides theory basis and instruction for conserving wild germplasm resources.
6 DATA AVAILABILITY STATEMENTAll data generated or analyzed during this study are included in this published article.
7 ACKNOWLEDGEMENTWe are very much indebted to DONG Dong, MA Lin, SUI Jixing, KOU Qi, GAN Zhibin, GONG Lin, WANG Jinbao, WANG Yueyun, XU Yong and SUN Yue for their work in the field and laboratory.
Alam M M M, de Croos M D S T, Pálsson S. 2017. Mitochondrial DNA variation reveals distinct lineages in Penaeus semisulcatus (Decapoda, Penaeidae) from the Indo-West Pacific Ocean. Marine Ecology, 38(2): e12406. DOI:10.1111/maec.12406 |
Alam M M M, Westfall K M, Pálsson S. 2015. Mitochondrial DNA variation reveals cryptic species in Fenneropenaeus indicus. Bulletin of Marine Science, 91(1): 15-31. |
Alam M M M, Westfall K M, Pálsson S. 2016. Mitogenomic variation of Bangladesh Penaeus monodon (Decapoda:Panaeidae) and reassessment of its phylogeography in the Indo-West Pacific region. Hydrobiologia, 763: 249-265. DOI:10.1007/s10750-015-2381-3 |
Avise J C. 2000. Phylogeography: The History and Formation of Species. Harvard University Press, Cambridge, Massachusetts, USA. 464p.
|
Bandelt H J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16(1): 37-48. DOI:10.1093/oxfordjournals.molbev.a026036 |
Cheang C C, Chu K H, Ang Jr P O. 2010. Phylogeography of the marine macroalga Sargassum hemiphyllum(Phaeophyceae, Heterokontophyta) in northwestern Pacific. Molecular Ecology, 19(14): 2 933-2 948. DOI:10.1111/mec.2010.19.issue-14 |
de Bruyn M, Nugroho E, Hossain M M, Wilson J C, Mather P B. 2005. Phylogeographic evidence for the existence of an ancient biogeographic barrier:the Isthmus of Kra Seaway. Heredity, 94(3): 370-378. DOI:10.1038/sj.hdy.6800613 |
Dong Y W, Wang H S, Han G D, Ke C H, Zhan X, Nakano T, Williams G A. 2012. The impact of Yangtze River discharge, ocean currents and historical events on the biogeographic pattern of Cellana toreuma along the China Coast. PLoS One, 7(4): e36178. DOI:10.1371/journal.pone.0036178 |
Dou Y G, Yang S Y, Liu Z X, Shi X F, Li J, Yu H, Berne S. 2012. Sr-Nd isotopic constraints on terrigenous sediment provenances and Kuroshio Current variability in the Okinawa Trough during the late Quaternary. Palaeogeography, Palaeoclimatology, Palaeoecology, 365-366: 38-47. DOI:10.1016/j.palaeo.2012.09.003 |
Du X W, Cai S S, Yu C G, Jiang X Q, Lin L S, Gao T X, Han Z Q. 2016. Population genetic structure of mantis shrimps Oratosquilla oratoria:testing the barrier effect of the Yangtze River outflow. Biochemical Systematics and Ecology, 66: 12-18. DOI:10.1016/j.bse.2016.02.033 |
Excoffier L, Lischer H E L. 2010. Arlequin suite ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10(3): 564-567. DOI:10.1111/men.2010.10.issue-3 |
Excoffier L, Smouse P E, Quattro J M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes:application to human mitochondrial DNA restriction data. Genetics, 131(2): 479-491. |
Feng B B, Li J L, Niu D H, Chen L, Zheng Y F, Zheng K H. 2008. Compared analysis on the sequences of Mitochondrial CR Gene and COI Gene Fragments of nine wild stocks of Portunus trituberculatus from the marginal seas of China. Chinese Journal of Zoology, 43(2): 28-36. |
Fu Y X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147(2): 915-925. |
Grant W A S, Bowen B W. 1998. Shallow population histories in deep evolutionary lineages of marine fishes:insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89(5): 415-426. DOI:10.1093/jhered/89.5.415 |
Hamano T, Matsuura S. 1987. Egg size, duration of incubation, and larval development of the Japanese mantis shrimp in the laboratory. Nippon Suisan Gakkaishi, 53(1): 23-39. DOI:10.2331/suisan.53.23 |
Han Z Q, Gao T X, Yanagimoto T, Sakurai Y. 2008. Genetic population structure of Nibea albiflora in Yellow Sea and East China Sea. Fisheries Science, 74(3): 544-552. DOI:10.1111/j.1444-2906.2008.01557.x |
Han Z Q, Zheng W, Chen G B, Shui B N, Liu S F, Zhuang Z M. 2015a. Population genetic structure and larval dispersal strategy of portunid crab Charybdis bimaculata in Yellow sea and East China sea. Mitochondrial DNA, 26(3): 402-408. DOI:10.3109/19401736.2013.840592 |
Han Z Q, Zhu W B, Zheng W, Li P F, Shui B N. 2015b. Significant genetic differentiation between the Yellow Sea and East China Sea populations of cocktail shrimp Trachypenaeus curvirostris revealed by the mitochondrial DNA COI gene. Biochemical Systematics and Ecology, 59: 78-84. DOI:10.1016/j.bse.2014.12.028 |
Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature, 405(6789): 907-913. DOI:10.1038/35016000 |
Hurwood D A, Dammannagoda S, Krosch M N, Jung H, Salin K R, Youssef M A B H, de Bruyn M, Mather P B. 2014. Impacts of climatic factors on evolution of molecular diversity and the natural distribution of wild stocks of the giant freshwater prawn (Macrobrachium rosenbergii). Freshwater Science, 33(1): 217-231. DOI:10.1086/675243 |
Hwang J S, Wong C K. 2005. The China Coastal Current as a driving force for transporting Calanus sinicus (Copepoda:Calanoida) from its population centers to waters off Taiwan and Hong Kong during the winter northeast monsoon period. Journal of Plankton Research, 27(2): 205-210. |
Ju Y M, Hsu C H, Fang L S, Lin H D, Wu J H, Han C C, Chen I S, Chiang T Y. 2013. Population structure and demographic history of Sicyopterus japonicus(Perciformes; Gobiidae) in Taiwan inferred from mitochondrial control region sequences. Genetics and Molecular Research, 12(3): 4 046-4 059. DOI:10.4238/2013.September.27.6 |
Kitamura A, Takano O, Takata H, Omote H. 2001. Late Pliocene-early Pleistocene paleoceanographic evolution of the Sea of Japan. Palaeogeography, Palaeoclimatology, Palaeoecology, 172(1-2): 81-98. DOI:10.1016/S0031-0182(01)00272-3 |
Kong L F, Li Q. 2009. Genetic evidence for the existence of cryptic species in an endangered clam Coelomactra antiquata. Marine Biology, 156(7): 1 507-1 515. DOI:10.1007/s00227-009-1190-5 |
Li N S, Zhao S L, Wasiliev B. 2000. Geology of Marginal Seas in the Northwest Pacific. Heilongjiang Education Press, Harbin, China. 535p. (in Chinese)
|
Librado P, Rozas J. 2009. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1 451-1 452. DOI:10.1093/bioinformatics/btp187 |
Liu H Y, Wang D X, Jiang Y S, Chen L. 2012. The effects of salinity on survival and food intake in mantis shrimp Oratosquilla oratoria larvae. Journal of Dalian Fisheries University, 27(4): 311-314. |
Liu H Y, Xu H L, Lin Y J. 2006. The effect of salinity on survival and growth of mantis shrimp (Oratosquilla oratoria) in Dalian coast. Journal of Dalian Fisheries University, 21(2): 180-183. |
Liu J X, Gao T X, Wu S F, Zhang Y P. 2007. Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus(Temminck & Schlegel, 1845). Molecular Ecology, 16(2): 275-288. |
Lu Z H, Xue L J, Zhang Y Z. 2013. Species composition and quantitative distribution of Stomatopod in the East China Sea. Journal of Natural Resources, 28(12): 2 159-2 168. |
Lui K K Y, Leung P T Y, Ng W C, Leung K M Y. 2010. Genetic variation of Oratosquilla oratoria (Crustacea:Stomatopoda) across Hong Kong waters elucidated by mitochondrial DNA control region sequences. Journal of the Marine Biological Association of the United Kingdom, 90(3): 623-631. DOI:10.1017/S0025315409990841 |
Nakajima Y, Matsuki Y, Lian C L, Fortes M D, Uy W H, Campos W L, Nakaoka M, Nadaoka K. 2014. The Kuroshio Current influences genetic diversity and population genetic structure of a tropical seagrass, Enhalus acoroides. Molecular Ecology, 23(24): 6 029-6 044. DOI:10.1111/mec.12996 |
Ni G, Li Q, Kong L F, Yu H. 2014. Comparative phylogeography in marginal seas of the northwestern Pacific. Molecular Ecology, 23(3): 534-548. DOI:10.1111/mec.12620 |
Ni G, Li Q, Kong L F, Zheng X D. 2012. Phylogeography of bivalve Cyclina sinensis:testing the historical glaciations and Changjiang River outflow hypotheses in Northwestern Pacific. PLoS One, 7(11): e49487. DOI:10.1371/journal.pone.0049487 |
Palumbi S R. 1994. Genetic divergence, reproductive isolation, and marine speciation. Annual Review of Ecology and Systematics, 25: 547-572. DOI:10.1146/annurev.es.25.110194.002555 |
Saitou N, Nei M. 1987. The neighbor-joining method:a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4): 406-425. |
Shen K N, Jamandre B W, Hsu C C, Tzeng W N, Durand J D. 2011. Plio-Pleistocene sea level and temperature fluctuations in the northwestern Pacific promoted speciation in the globally-distributed flathead mullet Mugil cephalus. BMC Evolutionary Biology, 11: 83. DOI:10.1186/1471-2148-11-83 |
Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123(3): 585-595. |
Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The CLUSTAL_X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24): 4 876-4 882. DOI:10.1093/nar/25.24.4876 |
Wang B, Zhang X L, Sun P X. 1998. On biological characters and artificial seedling rearing techniques of mantis shrimp(Oratosquilla oratoria). Journal of Oceanography of Huanghai & Bohai Seas, 16(2): 64-73. |
Wang C L, Xu S L, Mei W X, Zhang Y H, Wang D G. 1996. A biological basic character of Oratosquilla oratoria. Journal of Zhejiang College of Fisheries, 15(1): 60-62. |
Wang P X. 1999. Response of western Pacific marginal seas to glacial cycles:paleoceanographic and sedimentological features. Marine Geology, 156(1-4): 5-39. DOI:10.1016/S0025-3227(98)00172-8 |
Yang D Z. 2011. Numerical study on the Kuroshio intrusion on the East China Sea shelf and its influence on the biological process. Institute of Oceanology, Graduate University of Chinese Academy of Sciences, Qingdao, China. 108p. (in Chinese with English abstract)
|
Yasuda N, Nagai S, Hamaguchi M, Okaji K, Gérard K, Nadaoka K. 2009. Gene flow of Acanthaster planci (L.) in relation to ocean currents revealed by microsatellite analysis. Molecular Ecology, 18(8): 1 574-1 590. DOI:10.1111/mec.2009.18.issue-8 |
Zhang D Z, Ding G, Ge B M, Liu Q N, Zhang H B, Tang B P, Yang G. 2016. Geographical distribution, dispersal and genetic divergence of the mantis shrimp Oratosquilla oratoria (Stomatopoda:Squillidae) in China Sea. Biochemical Systematics and Ecology, 65: 1-8. DOI:10.1016/j.bse.2016.01.009 |
Zhang D Z, Ding G, Ge B M, Zhang H B, Tang B P, Yang G. 2014. Comparative phylogeography of two marine species of crustacean:recent divergence and expansion due to environmental changes. Gene, 550(1): 141-147. DOI:10.1016/j.gene.2014.08.006 |
Zhang D, Ding G, Ge B, Zhang H, Tang B. 2012. Population genetic structure and historical demography of Oratosquilla oratoria revealed by mitochondrial DNA sequences. Russian Journal of Genetics, 48(12): 1 232-1 238. DOI:10.1134/S1022795412110142 |
| Alam M M M, de Croos M D S T, Pálsson S, 2017. Mitochondrial DNA variation reveals distinct lineages in Penaeus semisulcatus (Decapoda, Penaeidae) from the Indo-West Pacific Ocean. Marine Ecology, 38(2): e12406. Doi: 10.1111/maec.12406 |
| Alam M M M, Westfall K M, Pálsson S, 2015. Mitochondrial DNA variation reveals cryptic species in Fenneropenaeus indicus. Bulletin of Marine Science, 91(1): 15–31. |
| Alam M M M, Westfall K M, Pálsson S, 2016. Mitogenomic variation of Bangladesh Penaeus monodon (Decapoda:Panaeidae) and reassessment of its phylogeography in the Indo-West Pacific region. Hydrobiologia, 763: 249–265. Doi: 10.1007/s10750-015-2381-3 |
| Avise J C. 2000. Phylogeography: The History and Formation of Species. Harvard University Press, Cambridge, Massachusetts, USA. 464p. |
| Bandelt H J, Forster P, Röhl A, 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16(1): 37–48. Doi: 10.1093/oxfordjournals.molbev.a026036 |
| Cheang C C, Chu K H, Ang Jr P O, 2010. Phylogeography of the marine macroalga Sargassum hemiphyllum(Phaeophyceae, Heterokontophyta) in northwestern Pacific. Molecular Ecology, 19(14): 2 933–2 948. Doi: 10.1111/mec.2010.19.issue-14 |
| de Bruyn M, Nugroho E, Hossain M M, Wilson J C, Mather P B, 2005. Phylogeographic evidence for the existence of an ancient biogeographic barrier:the Isthmus of Kra Seaway. Heredity, 94(3): 370–378. Doi: 10.1038/sj.hdy.6800613 |
| Dong Y W, Wang H S, Han G D, Ke C H, Zhan X, Nakano T, Williams G A, 2012. The impact of Yangtze River discharge, ocean currents and historical events on the biogeographic pattern of Cellana toreuma along the China Coast. PLoS One, 7(4): e36178. Doi: 10.1371/journal.pone.0036178 |
| Dou Y G, Yang S Y, Liu Z X, Shi X F, Li J, Yu H, Berne S, 2012. Sr-Nd isotopic constraints on terrigenous sediment provenances and Kuroshio Current variability in the Okinawa Trough during the late Quaternary. Palaeogeography, Palaeoclimatology, Palaeoecology, 365-366: 38–47. Doi: 10.1016/j.palaeo.2012.09.003 |
| Du X W, Cai S S, Yu C G, Jiang X Q, Lin L S, Gao T X, Han Z Q, 2016. Population genetic structure of mantis shrimps Oratosquilla oratoria:testing the barrier effect of the Yangtze River outflow. Biochemical Systematics and Ecology, 66: 12–18. Doi: 10.1016/j.bse.2016.02.033 |
| Excoffier L, Lischer H E L, 2010. Arlequin suite ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10(3): 564–567. Doi: 10.1111/men.2010.10.issue-3 |
| Excoffier L, Smouse P E, Quattro J M, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes:application to human mitochondrial DNA restriction data. Genetics, 131(2): 479–491. |
| Feng B B, Li J L, Niu D H, Chen L, Zheng Y F, Zheng K H, 2008. Compared analysis on the sequences of Mitochondrial CR Gene and COI Gene Fragments of nine wild stocks of Portunus trituberculatus from the marginal seas of China. Chinese Journal of Zoology, 43(2): 28–36. |
| Fu Y X, 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147(2): 915–925. |
| Grant W A S, Bowen B W, 1998. Shallow population histories in deep evolutionary lineages of marine fishes:insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89(5): 415–426. Doi: 10.1093/jhered/89.5.415 |
| Hamano T, Matsuura S, 1987. Egg size, duration of incubation, and larval development of the Japanese mantis shrimp in the laboratory. Nippon Suisan Gakkaishi, 53(1): 23–39. Doi: 10.2331/suisan.53.23 |
| Han Z Q, Gao T X, Yanagimoto T, Sakurai Y, 2008. Genetic population structure of Nibea albiflora in Yellow Sea and East China Sea. Fisheries Science, 74(3): 544–552. Doi: 10.1111/j.1444-2906.2008.01557.x |
| Han Z Q, Zheng W, Chen G B, Shui B N, Liu S F, Zhuang Z M, 2015a. Population genetic structure and larval dispersal strategy of portunid crab Charybdis bimaculata in Yellow sea and East China sea. Mitochondrial DNA, 26(3): 402–408. Doi: 10.3109/19401736.2013.840592 |
| Han Z Q, Zhu W B, Zheng W, Li P F, Shui B N, 2015b. Significant genetic differentiation between the Yellow Sea and East China Sea populations of cocktail shrimp Trachypenaeus curvirostris revealed by the mitochondrial DNA COI gene. Biochemical Systematics and Ecology, 59: 78–84. Doi: 10.1016/j.bse.2014.12.028 |
| Hewitt G, 2000. The genetic legacy of the Quaternary ice ages. Nature, 405(6789): 907–913. Doi: 10.1038/35016000 |
| Hurwood D A, Dammannagoda S, Krosch M N, Jung H, Salin K R, Youssef M A B H, de Bruyn M, Mather P B, 2014. Impacts of climatic factors on evolution of molecular diversity and the natural distribution of wild stocks of the giant freshwater prawn (Macrobrachium rosenbergii). Freshwater Science, 33(1): 217–231. Doi: 10.1086/675243 |
| Hwang J S, Wong C K, 2005. The China Coastal Current as a driving force for transporting Calanus sinicus (Copepoda:Calanoida) from its population centers to waters off Taiwan and Hong Kong during the winter northeast monsoon period. Journal of Plankton Research, 27(2): 205–210. |
| Ju Y M, Hsu C H, Fang L S, Lin H D, Wu J H, Han C C, Chen I S, Chiang T Y, 2013. Population structure and demographic history of Sicyopterus japonicus(Perciformes; Gobiidae) in Taiwan inferred from mitochondrial control region sequences. Genetics and Molecular Research, 12(3): 4 046–4 059. Doi: 10.4238/2013.September.27.6 |
| Kitamura A, Takano O, Takata H, Omote H, 2001. Late Pliocene-early Pleistocene paleoceanographic evolution of the Sea of Japan. Palaeogeography, Palaeoclimatology, Palaeoecology, 172(1-2): 81–98. Doi: 10.1016/S0031-0182(01)00272-3 |
| Kong L F, Li Q, 2009. Genetic evidence for the existence of cryptic species in an endangered clam Coelomactra antiquata. Marine Biology, 156(7): 1 507–1 515. Doi: 10.1007/s00227-009-1190-5 |
| Li N S, Zhao S L, Wasiliev B. 2000. Geology of Marginal Seas in the Northwest Pacific. Heilongjiang Education Press, Harbin, China. 535p. (in Chinese) |
| Librado P, Rozas J, 2009. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1 451–1 452. Doi: 10.1093/bioinformatics/btp187 |
| Liu H Y, Wang D X, Jiang Y S, Chen L, 2012. The effects of salinity on survival and food intake in mantis shrimp Oratosquilla oratoria larvae. Journal of Dalian Fisheries University, 27(4): 311–314. |
| Liu H Y, Xu H L, Lin Y J, 2006. The effect of salinity on survival and growth of mantis shrimp (Oratosquilla oratoria) in Dalian coast. Journal of Dalian Fisheries University, 21(2): 180–183. |
| Liu J X, Gao T X, Wu S F, Zhang Y P, 2007. Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus(Temminck & Schlegel, 1845). Molecular Ecology, 16(2): 275–288. |
| Lu Z H, Xue L J, Zhang Y Z, 2013. Species composition and quantitative distribution of Stomatopod in the East China Sea. Journal of Natural Resources, 28(12): 2 159–2 168. |
| Lui K K Y, Leung P T Y, Ng W C, Leung K M Y, 2010. Genetic variation of Oratosquilla oratoria (Crustacea:Stomatopoda) across Hong Kong waters elucidated by mitochondrial DNA control region sequences. Journal of the Marine Biological Association of the United Kingdom, 90(3): 623–631. Doi: 10.1017/S0025315409990841 |
| Nakajima Y, Matsuki Y, Lian C L, Fortes M D, Uy W H, Campos W L, Nakaoka M, Nadaoka K, 2014. The Kuroshio Current influences genetic diversity and population genetic structure of a tropical seagrass, Enhalus acoroides. Molecular Ecology, 23(24): 6 029–6 044. Doi: 10.1111/mec.12996 |
| Ni G, Li Q, Kong L F, Yu H, 2014. Comparative phylogeography in marginal seas of the northwestern Pacific. Molecular Ecology, 23(3): 534–548. Doi: 10.1111/mec.12620 |
| Ni G, Li Q, Kong L F, Zheng X D, 2012. Phylogeography of bivalve Cyclina sinensis:testing the historical glaciations and Changjiang River outflow hypotheses in Northwestern Pacific. PLoS One, 7(11): e49487. Doi: 10.1371/journal.pone.0049487 |
| Palumbi S R, 1994. Genetic divergence, reproductive isolation, and marine speciation. Annual Review of Ecology and Systematics, 25: 547–572. Doi: 10.1146/annurev.es.25.110194.002555 |
| Saitou N, Nei M, 1987. The neighbor-joining method:a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4): 406–425. |
| Shen K N, Jamandre B W, Hsu C C, Tzeng W N, Durand J D, 2011. Plio-Pleistocene sea level and temperature fluctuations in the northwestern Pacific promoted speciation in the globally-distributed flathead mullet Mugil cephalus. BMC Evolutionary Biology, 11: 83. Doi: 10.1186/1471-2148-11-83 |
| Tajima F, 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123(3): 585–595. |
| Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G, 1997. The CLUSTAL_X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24): 4 876–4 882. Doi: 10.1093/nar/25.24.4876 |
| Wang B, Zhang X L, Sun P X, 1998. On biological characters and artificial seedling rearing techniques of mantis shrimp(Oratosquilla oratoria). Journal of Oceanography of Huanghai & Bohai Seas, 16(2): 64–73. |
| Wang C L, Xu S L, Mei W X, Zhang Y H, Wang D G, 1996. A biological basic character of Oratosquilla oratoria. Journal of Zhejiang College of Fisheries, 15(1): 60–62. |
| Wang P X, 1999. Response of western Pacific marginal seas to glacial cycles:paleoceanographic and sedimentological features. Marine Geology, 156(1-4): 5–39. Doi: 10.1016/S0025-3227(98)00172-8 |
| Yang D Z. 2011. Numerical study on the Kuroshio intrusion on the East China Sea shelf and its influence on the biological process. Institute of Oceanology, Graduate University of Chinese Academy of Sciences, Qingdao, China. 108p. (in Chinese with English abstract) |
| Yasuda N, Nagai S, Hamaguchi M, Okaji K, Gérard K, Nadaoka K, 2009. Gene flow of Acanthaster planci (L.) in relation to ocean currents revealed by microsatellite analysis. Molecular Ecology, 18(8): 1 574–1 590. Doi: 10.1111/mec.2009.18.issue-8 |
| Zhang D Z, Ding G, Ge B M, Liu Q N, Zhang H B, Tang B P, Yang G, 2016. Geographical distribution, dispersal and genetic divergence of the mantis shrimp Oratosquilla oratoria (Stomatopoda:Squillidae) in China Sea. Biochemical Systematics and Ecology, 65: 1–8. Doi: 10.1016/j.bse.2016.01.009 |
| Zhang D Z, Ding G, Ge B M, Zhang H B, Tang B P, Yang G, 2014. Comparative phylogeography of two marine species of crustacean:recent divergence and expansion due to environmental changes. Gene, 550(1): 141–147. Doi: 10.1016/j.gene.2014.08.006 |
| Zhang D, Ding G, Ge B, Zhang H, Tang B, 2012. Population genetic structure and historical demography of Oratosquilla oratoria revealed by mitochondrial DNA sequences. Russian Journal of Genetics, 48(12): 1 232–1 238. Doi: 10.1134/S1022795412110142 |
 2018, Vol. 36
2018, Vol. 36