Institute of Oceanology, Chinese Academy of Sciences
Article Information
- ZOU Shanmei(邹山梅), FEI Cong(费聪), YANG Weinan(杨未男), HUANG Zheng(黄铮), HE Meilin(何梅琳), WANG Changhai(王长海)
- High-efficiency 18S microalgae barcoding by coalescent, distance and character-based approaches: a test in Chlorella and Scenedesmus
- Chinese Journal of Oceanology and Limnology, 36(5): 1771-1777
- http://dx.doi.org/10.1007/s00343-018-7201-y
Article History
- Received Jul. 20, 2017
- accepted in principle Aug. 26, 2017
- accepted for publication Oct. 24, 2017
Microalgae are diverse in aquatic habitats, and play a crucial role in the global ecosystem. Identifying microalgal biodiversity is significant for the use of microalgae in biofuel production, purification of wastewater, and extractions in high-added-value foods and pharmaceutical products. However, microalgae identification is extremely difficult because of their small size and the lack of obvious structural features among species.
DNA barcoding has been shown to be efficient in species identification (Hebert et al., 2003; Krawczyk et al., 2014), and the combination of multiple gene loci has been proved effective in DNA barcoding. The Plant Working Group of the Consortium for the Barcode of Life (CBOL) proposed the plastid genes rbcL (RuBisCO large subunit) and matK (megakaryocyte-associated tyrosine kinase) as standard barcodes for land plants (CBOL Plant Working Group, 2009). However, matk is absent in green algae. Several gene loci, such as rbcL, ITS and tufA (translation elongation factor 1-alpha), have often been used in the identification of green algae (Saunders and Kucera, 2010; Heeg and Wolf, 2015). Our previous studies also indicated that rbcL, ITS and tufA were useful for barcoding green algae (Zou et al., 2016a, b). 18S, which has relatively conserved sequences, is often used in phylogenetic analysis of plants and animals. It is also employed in microalgae identification because of the complex diversity of microalgae. Whether 18S can really help in distinguishing microalgae merits evaluation.
Monophyly and distance-based barcoding, originally proposed by Hebert et al. (2003), are both controversial. Recently, the use of coalescent and character-based barcoding has been proposed. Complementary barcoding analyses such as the P ID (Liberal) method of species delimitation, GMYC (Fujisawa and Barracloug, 2013), the PTP model, Automatic Barcode Gap Discovery (ABGD) (Kekkonen and Hebert, 2014) and character-based approaches (Rach et al., 2008) have also been proposed. The GMYC model sets a threshold to delineate evolutionary significant units (ESUs) akin to species. The P ID (Liberal) method of species delimitation allows differing species boundary hypotheses to be investigated by enabling the user a priori to assign taxa to putative species groups on a phylogenetic tree. The PTP model distinguishes specimens in both populations and at species level using coalescence theory. The ABGD method can assign the sequences into potential species based on the barcode gap whenever the divergence within the same species is smaller than that among organisms from different species. The character-based barcoding approach is based on the concept that members of a given taxonomic group have the same diagnostic characters that are absent from comparable groups. This has the logical advantage that it will fail to diagnose the specimens when diagnostic character data are lacking, in comparison with using distance. Thus, a combination of distance, coalescent and character-based approaches may be effective in testing the barcoding efficiency of 18S in microalgae.
The microalgae genera Chlorella and Scenedesmus are both well known worldwide, and may be ideal for producing biofuel owing to the substantial amounts of lipids, proteins and carbohydrates they contain. Previous studies, including our own, show that rbcL, ITS and tufA may assist in barcoding Chlorella and Scenedesmus samples (Krienitz et al., 2004; Luo et al., 2006, 2010; Škaloud et al., 2014; Heeg and Wolf, 2015; Zou et al., 2016a, b). In this paper, the same Chlorella and Scenedesmus samples collected in our previous study (Zou et al., 2016a, b) are employed again to test the real efficiency of 18S for barcoding microalgae with coalescent (GMYC, P ID and PTP), distance (ABGD) and character-based analysis. We also compare our findings with the rbcL, ITS and tufA barcoding results in Zou et al.(2016a, b).
2 MATERIAL AND METHOD 2.1 Algal sampling and culturingA total of 56 Chlorella and 54 Scenedesmus samples collected in Zou et al.(2016a, b), which were collected from marine, freshwater, Arctic and terrestrial areas, were analyzed (Table S1) (Zou et al., 2016a, b). Table S1 details the taxa studied.
2.2 DNA extraction, amplification and sequencingDNA was extracted using the Qiagen DNEasy Plant Extraction kit (Qiagen Inc., Valencia, CA, USA). Primers for amplifying and sequencing 18S barcode region were: 18SF: AAC CTG GTT GAT CCT GCC AGT; 18SR: TTG ATC CTT CTG CAG GTT CAC C; and 528R: TGC CAG CAG CYG CGG TAA TTC CAG C (Bock et al., 2011). PCR reactions were carried out in a total volume of 25 μL, using 2× DreamTaq Green PCR Master Mix (US). The PCR products were sent to the Beijing Genomics Institute (BGI) for bidirectional sequencing.
2.3 Sequence alignment and phylogenetic reconstructionForward and reverse sequences of 18S were assembled and edited in Sequencher (Gene Codes Corporation) and aligned by MAFFT 6.717 (https://mafft.cbrc.jp/alignment/software/). Construction of the Neighbor joining tree was based on the Kimura 2-parameter (K2P) distance model (Hebert et al., 2003) in MEGA 7.0 (Kumar et al., 2016). The jModeltest v.0.1.1 (Posada, 2008) was used to estimate the best substitution model for Bayesian analysis. The most appropriate model was GTR+G. The Bayesian analyses were conducted in BEAST (Drummond and Rambaut, 2007).
2.4 GMYC, PTP and P ID species boundary delimitationFor GMYC analysis, firstly, a linearized Bayesian phylogenetic tree was calculated employing a Yule pure birth model tree prior using BEAST (Drummond and Rambaut, 2007). Substitution models, empirical base frequencies, four gamma categories, all codon positions partitioned with unlinked base frequencies and substitution rates were set in BEAUTi v. 1.7.1. The length of MCMC chain was 40 000 000 sampling every 4 000. Log-Combiner v1.7.1 was used to merge two independent runs with 20% burn-in. Maximum clade credibility trees with a 0.5 posterior probability limit, and node heights of target tree, were constructed in TreeAnnotator v1.7.1. R (R Core Team, 2014) and used to perform single-threshold GMYC analyses using the APE and SPLITS packages.
The species delimitation plugin within Geneious Pro v5.5.4 (Biomatters; http://www.geneious.com) was investigated to assess species boundary hypotheses across the Bayesian gene tree. Maximumlikelihood trees were inferred by employing PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/).
The PTP method was implemented in a web server (http://species.h-its.org/).
2.5 ABGDThe ABGD method is available at http://www.abi.snv.jussieu.fr/public/abgd/. The 18S sequence data were processed in ABGD using the K2P nucleotide substitution model. The maximum value of intraspecific divergence was set in advance at between 0.001 and 0.1. The gap width was set at 1.0.
2.6 Character-based DNA barcode analysesThe character-based barcoding analysis was conducted in the characteristic attribute organization system (CAOS) system, which comprises two programs: P-Gnome and P-Elf (Rach et al., 2008; Bergmann et al., 2009). The guide tree of 18S was produced using the programs PAUP v4.0b10, which were then incorporated into a NEXUS file containing the sequence data in MacClade (Maddison and Maddision, 2001). Then the incorporated NEXUS data set was conducted in CAOS system where the p-gnome script was used to identify characters.
3 RESULTA total of 110 18S sequences of Chlorella and Scenedesmus samples were analyzed in this study (Table S1). The newly obtained 18S sequences from this study were submitted to the National Center for Biotechnology Information (NCBI) with the accession numbers KX494985–KX495094. The 18S sequence had a length of 938 bp to 1 680 bp.
3.1 Coalescent barcoding analysisGenerally, the traditional barcoding analysis of NJ and Bayesian trees showed consistent topology, which separated some species, e.g. C. saccharophila and S. quadricauda (Fig. 1). Several species (e.g. C. sorokiniana, C. vulgaris; and S. bajacalifornicus) that were divided into two clades, respectively, may potentially be cryptic species. This result was consistent with our previous results that C. sorokiniana, C. vulgaris and S. bajacalifornicus were recovered as cryptic species (Zou et al., 2016a, b). Only a few species were revealed as paraphyletic clades, e.g. S. deserticola, S. armatus, S. bajacalifornicus, C. vulgaris Ⅱ and C. sorokiniana Ⅱ. The P ID analysis recovered most species as separate clades except C. vulgaris Ⅱ, S. bajacalifornicus Ⅰ and Ⅱ, and S. armatus, which were revealed as paraphyletic clades. All delimited species of 18S sequences possessed a P ID (Liberal) value > 0.5 except for two clades (Table S1).
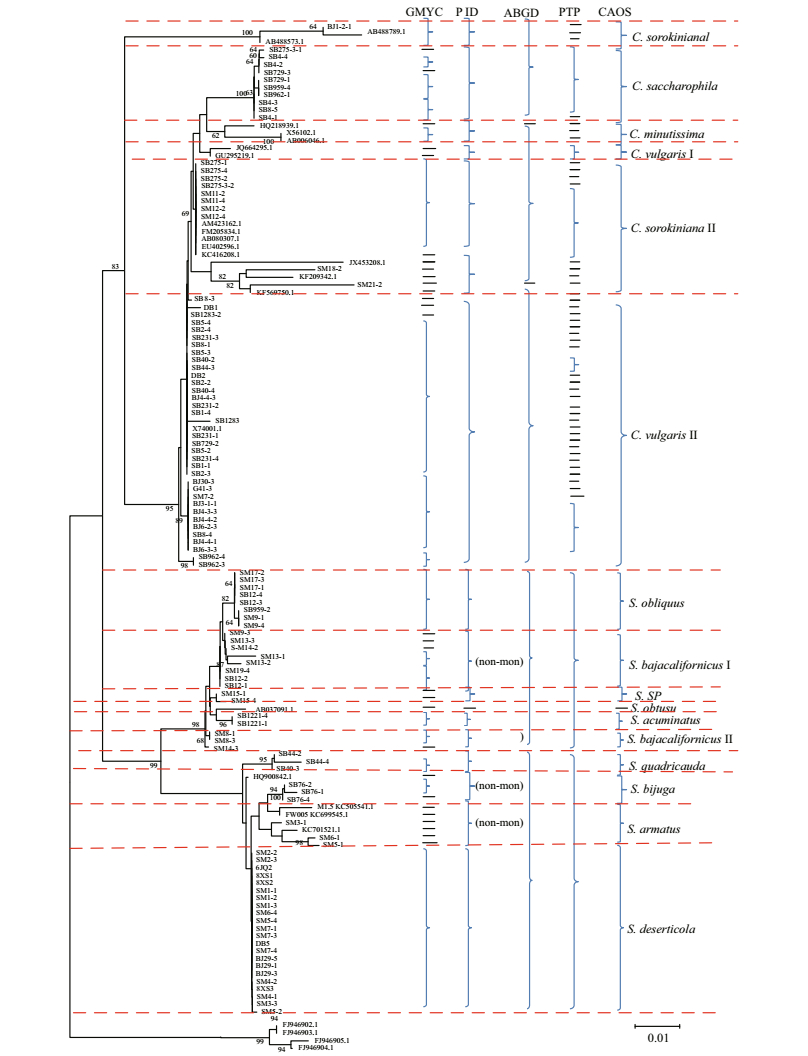
|
| Figure 1 Bayesian phylogenetic tree for the 18S gene Vertical braces on the right indicate the clades detected by approaches of the tree-based GMYC, PID and PTP; the distance-based ABGD; the character-based CAOS; and the final assignment. |
It was indicated that some specimens analyzed were over-splitted by the GMYC model (Fig. 1). The results of single-threshold analysis for 18S suggested 46 groups. The resolution produced by bPTP analysis was similar to the GYMC model. PTP analysis could distinguish most species except some (e.g. C. vulgaris Ⅱ) that were over-splitted (Fig. 1). Maximumlikelihood identification produced better resolution than Bayesian identification. In total, the 18S sequence recognized more than 30 independent entities by bPTP (Fig. 1).
3.2 ABGD analysisDifferent groups as candidate species were produced using the distance-based approach as implemented in the software ABGD. In total, the ABGD analysis revealed nine genetic groups when using restrictive values with an a priori genetic distance threshold of 0.77% (Fig. 1 and Fig.S1). It was apparent that the ABGD analysis revealed fewer genetic groups than other barcoding methods (Fig. 1).
3.3 Character-based barcodingBased on morphological identification, phylogenetic construction, and GMYC, PTP, P ID and ABGD analysis, the originally defined Chlorella and Scenedesmus clades (Fig. 1) were analyzed by searching for diagnostic characters using a characterbased barcoding approach. It was shown that most species were clearly distinguished in the characterbased DNA barcoding with more than three character attributes in 33 positions (Table 1). Only C. sorokiniana Ⅱ and C. vulgaris Ⅱ, and S. quadricauda and S. bijuga shared the same nucleotide characters. S. acuminatus and S. bajacalifornicus shared just one character attribute (T/C) (Table 1).

|
In recent years, DNA barcoding technology has been proved effective for species identification in both plant and animal kingdoms (Hebert et al., 2003; Krawczyk et al., 2014). It also offers a good opportunity to reveal microalgae diversity. A fragment of the COI gene has been proved useful as a universal marker for a "DNA barcode" for the identification of animals, owing to universal primers and variation in sequences (Hebert et al., 2003). However, in plants, the amplification of COI is often difficult and it is not varied enough to distinguish most species. While "rbcL+matk" is proposed as a core barcode for plants, the standard barcode for microalgae is ambiguous (China Plant BOL Group et al., 2011). Our previous studies have already shown that rbcL, tufA and ITS are effective in barcoding the microalgae Chlorella and Scenedesmus (Zou et al., 2016a, b). We have also proved that morphological identification of most Chlorella and Scenedesmus species is consistent with the barcoding results. Compared with the COI, rbcL, 16S and tufA genes, 18S sequences are relatively conserved. 18S is often used in the phylogeny of microalgae, and in some cases is used for microalgae identification. It is necessary to confirm the efficiency of 18S for identifying microalgae species. Using the same Chlorella and Scenedesmus samples as in Zou et al.(2016a, b), this study aimed to test the efficiency of 18S for barcoding green microalgae based on coalescent (GMYC, P ID and PTP), distance (ABGD) and character-based approaches. The results of 18S barcoding are compared with the rbcL, tufA and ITS results in Zou et al.(2016a, b).
Barcoding methods continue to develop, but potential limitations mean that no one method can distinguish all species. Combining multiple barcoding approaches can produce better resolution. While all barcoding methods produce consistent result the species identification could be determined. Although multiple barcoding methods produce inconsistent identification, the result produced by the approach that shows more advantages could be more accurate. Many studies have indicated that character-based approaches showed more advantages compared with other methods (DeSalle et al., 2005; Rach et al., 2008; Damm et al., 2010; Yassin et al., 2010; Zou et al., 2011, 2016a, b).
This study found that, in general, the GMYC and PTP analyses generated more genetic groups. Some species were over-splitted by GMYC and PTP analysis, which is consistent with previous results that the GMYC and PTP models often over-split some taxa (Talavera et al., 2013; Kekkonen and Hebert, 2014; Zou et al., 2016a, b). The ABGD-based approach identified most species as monophyletic clades. Another method of P ID identification, which was proved to be effective for reevaluating the treebased hypotheses for species hypothesis, also recovered many species as monophyletic clades. In our previous study, the P ID method separated all Chlorella and Scenedesmus species as monophyletic clades by rbcL, ITS and tufA barcoding sequences (Zou, 2016a, b). Finally, in previous studies, the character-based analysis of rbcL, ITS, tufA and 16S could always distinguish the species with many character attributes, and the character-based method was proved the most effective barcoding approach (Zou et al., 2016a, b). In this study, with the exception of several lineages, all species could also be separated in character-based barcoding with more than three character attributes, which is consistent with previous results. Most species which could be identified by rbcL, ITS and tufA in our previous study (Zou et al., 2016a, b) could also be distinguished here by coalescent (GMYC, P ID and PTP), distance (ABGD) and character-based barcoding approaches by 18S. Thus, the results indicated that most Chlorella and Scenedesmus samples could be separated by barcoding 18S sequences.
In general, in comparison with previous barcoding results of rbcL, ITS and tufA, our study indicated that 18S showed a good ability to discriminate green microalgae taxa at species level using a combination of coalescent, distance and character-based barcoding approaches. However, it should be acknowledged here that Scenedesmus and Chlorella represent only a limited example of green microalgae samples. More microalgae genera should be collected to test 18S barcoding efficiency in future research.
5 CONCLUSIONIn this study, the efficiency of 18S for barcoding green microalgae was first tested by multiple approaches of coalescent, distance and characterbased barcoding. In comparison with barcoding results for rbcL, tufA and ITS in a previous study, we showed that 18S is highly efficient in barcoding Scenedesmus and Chlorella samples at species level by a combination of GMYC, PTP, P ID, ABGD and character-based barcoding approaches. Combining 18S with other gene markers for barcoding green microalgae, based on a combination of multiple barcoding approaches, may be a better choice.
6 ACKNOWLEDGEMENTWe acknowledge the collection of samples referred to in Zou et al.(2016a, b).
7 DATA AVAILABILITY STATEMENTThe data sets analyzed during the current study are in the National Center for Biotechnology Information (NCBI) (accession numbers KX494985–KX495094). They are not currently publicly available owing to the submission process of the National Center for Biotechnology Information (NCBI), but are available from the corresponding author on reasonable request.
Electronic supplementary materialSupplementary material (Supplementary Table S1 and Fig.S1) is available in the online version of this article at https://doi.org/10.1007/s00343-018-7201-y.
Bergmann T, Hadrys H, Breves G, Schierwater B. 2009. Character-based DNA barcoding:a superior tool for species classification. Berl. Munch. Tierarztl. Wochenschr., 122(11-12): 446-450.
|
Bock C, Pröschold T, Krienitz L. 2011. Updating the genus Dictyosphaerium and description of Mucidosphaerium gen. Nov. (Trebouxiophyceae) based on morphological and molecular data. J. Phycol, 47(3): 638-652.
DOI:10.1111/jpy.2011.47.issue-3 |
CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A., 106(31): 12 794-12 797.
DOI:10.1073/pnas.0905845106 |
China Plant BOL Group, Li D Z, Gao L M, Li H T, Wang H, Ge X J, Liu J Q, Chen Z D, Zhou S L, Chen S L, Yang J B, Fu C X, Zeng C X, Yan H F, Zhu Y J, Sun Y S, Chen S Y, Zhao L, Wang K, Yang T, Duan G W. 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A., 108(49): 19 641-19 646.
DOI:10.1073/pnas.1104551108 |
Damm S, Schierwater B, Hadrys H. 2010. An integrative approach to species discovery in odonates:from characterbased DNA barcoding to ecology. Mol. Ecol., 19(18): 3 881-3 893.
DOI:10.1111/j.1365-294X.2010.04720.x |
DeSalle R, Egan M G, Siddall M. 2005. The unholy trinity: taxonomy, species delimitation and DNA barcoding. Philos. Trans. Roy. Soc. B:Biol. Sci., 360(1462): 1 905-1 916.
DOI:10.1098/rstb.2005.1722 |
Drummond A J, Rambaut A. 2007. BEAST:bayesian evolutionary analysis by sampling trees. BMC Evol. Biol., 7: 214.
DOI:10.1186/1471-2148-7-214 |
Fujisawa T, Barraclough T G. 2013. Delimiting species using single-locus data and the generalized mixed Yule coalescent approach:a revised method and evaluation on No.5 ZOU et al.:High-efficiency 18S barcoding for green microalgae 1777 simulated data sets. Syst. Biol., 62(5): 707-724.
DOI:10.1093/sysbio/syt033 |
Hebert P D N, Cywinska A C, Ball S L, deWaard J R. 2003. Biological identifications through DNA barcodes. Philos. Trans. Roy. Soc. B:Biol. Sci., 270(1512): 313-321.
DOI:10.1098/rspb.2002.2218 |
Heeg J S, Wolf M. 2015. ITS2 and 18S rDNA sequencestructure phylogeny of Chlorella and allies (Chlorophyta, Trebouxiophyceae, Chlorellaceae). Plant Gene, 4: 20-28.
DOI:10.1016/j.plgene.2015.08.001 |
Kekkonen M, Hebert P D N. 2014. DNA barcode-based delineation of putative species:efficient start for taxonomic workflows. Mol. Ecol. Res., 14(4): 706-715.
DOI:10.1111/men.2014.14.issue-4 |
Krawczyk K, Szczecińska M, Sawicki J. 2014. Evaluation of 11 single-locus and seven multilocus DNA barcodes in Lamium L. (Lamiaceae). Mol. Ecol. Res., 14(2): 272-285.
DOI:10.1111/1755-0998.12175 |
Krienitz L, Hegewald E H, Hepperle D, Huss V A R, Rohrs T, Wolf M. 2004. Phylogenetic relationship of Chlorella and Parachlorellagen. nov. (Chlorophyta, Trebouxiophyceae). Phycologia, 43: 529-542.
DOI:10.2216/i0031-8884-43-5-529.1 |
Kumar S, Stecher G, Tamura K. 2016. MEGA7:Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol., 33(7): 1870-4.
DOI:10.1093/molbev/msw054 |
Luo W, Pflugmacher S, Pröeschold T, Walz N, Krienitz L. 2006. Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist, 157(3): 315-333.
DOI:10.1016/j.protis.2006.05.006 |
Luo W, Pröeschold T, Bock C, Krienitz L. 2010. Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biol., 12(3): 545-553.
DOI:10.1111/plb.2010.12.issue-3 |
Maddison D R, Maddison W P. 2001. MacClade: Analysis of Phylogeny and Character Evolution, Version 4.03. Sinauer Associates, Sunderland, MA. http://citeseer.ist.psu.edu/showciting?cid=922278
|
Posada D. 2008. jModelTest:phylogenetic model averaging. Mol. Biol. Evol., 25(7): 1 253-1 256.
DOI:10.1093/molbev/msn083 |
R Core Team. 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.
|
Rach J, DeSalle R, Sarkar I N, Schierwater B, Hadrys H. 2008. Character-based DNA barcoding allows discrimination of genera, species and populations in Odonata. Philos. Trans. Roy. Soc. B:Biol. Sci., 275(1632): 237-247.
DOI:10.1098/rspb.2007.1290 |
Saunders G W, Kucera H. 2010. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie Algol., 31(4): 487-528.
|
Škaloud P, Němcová Y, Pytela J, Bogdanov N I, Bock C, Pickinpaugh S H. 2014. Planktochlorella nurekis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel coccoid green alga carrying significant biotechnological potential. Fottea, 14(1): 53-62.
DOI:10.5507/fot.2014.004 |
Talavera G, Dincă V, Vila R. 2013. Factors affecting species delimitations with the GMYC model:insights from a butterfly survey. Methods Ecol. Evol., 4(12): 1 101-1 110.
DOI:10.1111/mee3.2013.4.issue-12 |
Yassin A, Markow T A, Narechania A, O'Grady P M, DeSalle R. 2010. The genus Drosophila as a model for testing tree-and character-based methods of species identification using DNA barcoding. Mol. Phylogenet. Evol., 57(2): 509-517.
DOI:10.1016/j.ympev.2010.08.020 |
Zou S M, Fei C, Song J M, Bao Y C, He M L, Wang C H. 2016a. Combining and comparing coalescent, distance and character-based approaches for barcoding microalgaes:a test with Chlorella-like species (Chlorophyta). PLoS One, 11(4): e015383.
|
Zou S M, Fei C, Wang C, Gao Z, Bao Y C, He M L, Wang C H. 2016b. How DNA barcoding can be more effective in microalgae identification:a case of cryptic diversity revelation in Scenedesmus (Chlorophyceae). Sci. Rep., 6: 36 822.
DOI:10.1038/srep36822 |
Zou S M, Li Q, Kong L F, Yu H, Zheng X D. 2011. Comparing the usefulness of distance, monophyly and characterbased DNA barcoding methods in species identification:a case study of Neogastropoda. PLoS One, 6(10): e26619.
DOI:10.1371/journal.pone.0026619 |
 2018, Vol. 36
2018, Vol. 36


