Institute of Oceanology, Chinese Academy of Sciences
Article Information
- SHAO Ting(邵婷), QIN Chuanjie(覃川杰), DUAN Huiguo(段辉国), YUAN Dengyue(袁登越), WEN Zhengyong(文正勇), WANG Jun(王钧), GE Fanglan(葛芳兰)
- Effects of feeding time on complement component C7 expression in Pelteobagrus vachellii subject to bacterial challenge
- Chinese Journal of Oceanology and Limnology, 36(6): 2358-2367
- http://dx.doi.org/10.1007/s00343-019-7270-6
Article History
- Received Oct. 11, 2017
- accepted in principle Nov. 19, 2017
- accepted for publication Dec. 11, 2017
2 College of Life Science, Neijiang Normal University, Key Laboratory of Sichuan Province for Fishes Conservation and Utilization in the Upper Reaches of the Yangtze River, Neijiang 641000, China
The complement system has been found in both vertebrates and a wide range of invertebrate species (Fujito et al., 2010; Wang et al., 2013; Liu et al., 2016). Complement was a system of plasma proteins, which were central to many defense mechanisms, and included more than 30 distinct plasma and membraneassociated proteins (Morgan et al., 2005; Wang et al., 2015). It was fundamental for innate immunity and a key modulator of specific immunity. The complement system mediates various immune effector functions, including the energization of inflammatory responses, the regulation of specific immune responses, and the killing and elimination of pathogens (Sun et al., 2013). These studies indicated that complement genes could play an essential role to clear pathogens.
Complement component, C7 was an important plasma protein, which involved in the cytolytic phase of complement activation, through a series of catalytic reactions with other terminal complement components. C7 played an essential role in the assembly of the cytolytically active membrane attack complex (MAC) within target cell membranes (Bossi et al., 2009); this was generated by the sequential addition of single molecules of C5b, C6-8, and variable numbers of C9 molecules as terminal complement components (TCCs). The invaded bacteria were cleared by the final MAC assembly with disrupting their membranes (Muller-Eberhard, 1986; Katagiri et al., 1999). Recently, the C7 gene had been identified in fish, including large yellow croaker Larimichthys crocea, miiuy croaker Miichthys miiuy, grass carp Ctenopharyngodon idella, and rock bream Oplegnathus fasciatus (Shen et al., 2012; Niroshana Wickramaarachchi et al., 2013; Wang et al., 2015; Guo et al., 2016). Due to stimulation by pathogenic bacteria or lipopolysaccharides (LPS), significant increase in C7 mRNA expression was observed in the above fish; this indicated that C7 participated in the innate immune responses of teleost fish to pathogenic infections (Shen et al., 2012; Niroshana Wickramaarachchi et al., 2013; Wang et al., 2015; Guo et al., 2016).
In animals, many studies have shown that shifting feeding time could disturb the synchrony of behavioral and physiological rhythms, which may lead to manifestations of the metabolic syndrome. Mukherji et al. (2015) reported that shifting the feeding time of mice to their rest phase increased the free fatty acid and glucagon levels, and created further metabolic alterations. In addition, altering mealtime affected daily cycles in the metabolic (glucose) and endocrine (cortisol and thyroid hormones) systems, digestive enzymes in gilthead seabream Sparus aurata (Montoya et al., 2010a). Working during the night had similar effects to shifting feeding times, likely due to disruption of the circadian rhythm, which could affect many physiological activities and immune function (Logan and Sarkar, 2012). Clock mutation can also suppress daily rhythms of circulating leukocytes and a greats of immune-related genes (Oishi et al., 2006). Weekly phase-shifts (6 h/week for 4 weeks) resulted in a significant difference of immune responses to high-dose (12.5 mg/kg) lipopolysaccharide (LPS) challenges in mice (Phillips et al., 2015). The disruption of circadian rhythms, from changes in feeding times or daily activities, may result in chronic illnesses, such as metabolic syndrome and cancer (Kalsbeek et al., 2011). However, there was little knowledge about the effects of shifting feeding time on immune molecules such as C7 in fish.
Darkbarbel catfish (Pelteobagrus vachellii) is an important commercial carnivorous aquaculture fish species in China (Li, 2010). Due to its high market value, the demand for this species has grown considerably in recent years, with total production reaching 417 347 tons in 2016 (Fisheries and Fisheries Administration Bureau of the Ministry of Agriculture, 2017). Previous studies indicated that juveniles of a similar species, Pelteobagrus fulvidraco, showed nocturnal feeding behavior (Yang et al., 2006). In our earlier study, the CLOCK gene expression in the brain, liver and intestine of P. vachellii was found to be under the control of physiological rhythms, and the acrophases of CLOCK mRNA expression in the brain, liver and intestine were at Zeitgeber time (ZT), of 21:35, 23:00 and 23:23, respectively (Qin and Shao, 2015). So, feeding at night may be better for P. vachellii, due to the synchronization with its feeding rhythms. However, farmed P. vachellii are generally fed twice per day, during daylight. Therefore, this study aimed to clone the C7 gene and analyze its expression in fish fed either during the day or night.
2 MATERIAL AND METHOD 2.1 Fish and experimental treatmentsA total of 500 fish (initial weight 12.32±4.71 g) were obtained from the local fish farm (Meishan, Sichuan, China), and acclimatized at 25±2℃ for 20 days in a flow-through system. Then, 360 healthy P. vachellii were randomly assigned to triplicate tanks of four treatment groups (30 fish/tank; 300 L/tank) (Fig. 1). Two groups were fed once a day in the daytime (08:00; daytime group), and two other groups were fed at night time (20:00; nighttime group), for 60 days with an automatic timer-feeder (EHEIM, model 3581, Germany). The diet was formulated to meet or exceed the known nutrient requirements of darkbarbel catfish; the 3-mm floating pellets (Neijiang Zhengda Inc., Sichuan, China) contained approximately 40% protein, 3.5% fat, 15% ash, 2% lysine and 12.5% moisture. The dissolved oxygen levels in each tank were approximately at saturation, due to continuous aeration. The fish were exposed to natural a light-dark cycle, with approximately 12-h light/12-h dark (from 7:00 to 7:00), and a natural water temperature (25±2℃). The ammonia and nitrate concentrations were measured once a week, and were always < 0.05 mg/L.
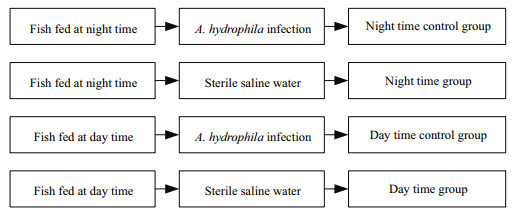
|
| Fig.1 Schematic diagram of the experimental design |
At the end of the feeding trial, four fish from each tank (twelve fish for each group) were anesthetized with MS-222 at 20:00. They were dissected and separately collect the liver, spleen, head kidney tissues and intestine, which were immediately stored in liquid nitrogen. In addition, for tissues expression analysis, ten tissue samples (brain, muscle, liver, head kidney, skin, spleen, heart, gill, intestine and fins) were separately collected from nine acclimatized healthy fish and immediately stored in liquid nitrogen.
2.2 Bacterial challenge assayTo examine the effect of feeding time on the Pv-C7 expression in P. vachellii, the fish remaining from the feeding trial (Section 2.1) were given an intraperitoneal injection with 100 μL live Gram-negative Aeromonas hydrophila (isolated from sick darkbarbel catfish and identified with 16s Rrna method) in saline water (8.5×109 CFU /mL per fish) (Qin et al., 2017). Thus, there were two groups in the bacterial challenge: fish fed at the daytime (08:00), and those fed at night (20:00). The other groups were the negative control groups. There were two groups in the negative control: fish fed at the daytime (08:00), and fish fed at night (20:00). The two negative groups were injected with 100 μL sterile saline water per fish. Details of the four treatments were shown in Fig. 1. Injections were carried out under anesthesia using MS-22 at 10 mg/L. Nine fish were randomly sampled at 0, 3, 12, 24, 48 and 96 h post-injection. The liver, head kidney, and spleen tissues were collected from each fish for total RNA isolation and further analysis.
2.3 RNA extraction and cloning of the C7 cDNAIn order to identify the Pv-C7 genes, C7 partial sequences from the liver transcriptome of P. vachellii were selected for further amplification (unpublished data) using local BLAST software (NCBI, USA). Following the protocol of RNAiso Plus kit (TaKaRa, Dalian, China), total RNA was extracted from the liver of P. vachellii. The full length C7 cDNA of P. vachellii was gained with the reverse transcription PCR (RT-PCR) and rapid amplification of cDNA ends (RACE) methods. For 3′-RACE and 5′-RACE, the first strand cDNA synthesis was carried out using SMARTScribe Reverse Transcriptase (Clontech, USA) with 3′-RACE CDS Primer and 5′-CDS primer A, respectively.
The SMART RACE cDNA Amplification Kit (Clontech, USA) was used to obtain the full length of the Pv-C7 gene with the manufacturer's instructions. For 5′-RACE, the primer set consisted of 5′-RACE GSP1 (5′-TGCTCATCTAAACCATCCTCC-3′) and the universal primer mix (UPM). For 3′-RACE, the primer set consisted of 3′-RACE GSP2 (5′-CTCACCCTTGAAGCAGACG-3′) and the UPM. The PCR reaction conditions defined in the Advantage TM 2 PAC kit (Clontech, USA) were carried out.
2.4 Cloning, sequencing and analysesThe purification of the PCR fragments and clone of cDNA fragments were carried out with electrophoresis on 1.5% agarose gel pGEM-T Easy vector, following the manufacturer's instructions (Promega Corporation, Madison, WI, USA). Recombinant bacteria were selected by blue/white method and identified by PCR.
The insert cDNA fragments in Plasmids were purified with miniprep kit (Promega), and the DNA sequencing was completed by Sangong (Sangon Biotech (Shanghai) Co., Ltd.) used. The Expert Protein Analysis System (http://us.expasy.org/) was use to analyze the C7 deduced amino acid sequence of P. vachellii. The BLAST (NCBI; http://ncbi.nlm.nih.gov/blast/) was used to perform similarity searches. Multiple alignment of the Pv-C7 was carried out with Clustal W Multiple Alignment.
2.5 Expression of the Pv-C7 gene in tissuesThe expression of P. vachellii C7 mRNA in tissues was detected by real-time RT-PCR analysis. Total RNA was also isolated with RNAiso Plus kit (Takara, Dalian, China). PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) was used to synthesize the first strand cDNA. The gene-specific primers QC7-S (5′-AATGAACTGAAGCGGGACA-3′) and QC7-A (5′-GCAGGATTGAGCGGTAAGC-3′) were designed to amplify the 172 bp Pv-C7 transcript. The primers of β-actin F (5′-CACTGTGCCCATCTACGAG-3′) and β-actin R (5′-CCATCTCCTGCTCGAAGTC-3′) were amplified the 200 bp fragment as a reference gene (Zheng et al., 2010). The PCR profiles were followed the instructions in the SYBR Premix Ex Taq (TaKaRa), and were performed on a Roche Nano Real-Time PCR System (Roche, USA). The 2-ΔΔCt method was used to calculate the gene mRNA relative expression levels (Livak and Schmittgen, 2001).
2.6 Statistical analysisThe differences in gene expression within tissues distribution and tissues from fish subject to the bacterial challenge assay were analyzed by one-way analysis of variance (ANOVA) and multiple comparison (Duncan) tests using SPSS software (SPSS v. 18.0). The difference expression of Pv-C7 mRNA at the same time point within A. hydrophila infection after shifting feeding time experiment were analyzed by independent-sample t-tests (SPSS v. 18.0), and P < 0.05 was considered to be statistically significant.
3 RESULT 3.1 C7 cDNA sequence of P. vachelliiA unigene was identified from the transcriptome of the P. vachellii liver tissue (BioProject ID: PRJNA362523); the fragments showed high identity with the C7 mRNA of C. idella (JN655169.1), Marsupenaeus japonicus (AB108542.1) and Homarus gammarus (AJ697860.1) in the GenBank database. The C7 cDNA of P. vachellii consisted of 2 647 bp, including a 5′ UTR of 52 bp, an open reading frame (ORF) of 2 454 bp, and a 3′ UTR of 141 bp. A consensus polyadenylation signal (AATAAA) sequence was observed in the 3′ UTR. The ORF encoded a protein consisting of 818 amino acid residues. The nucleotide and the deduced amino acid sequences of P. vachellii Pv-C7 cDNA were submitted to the GenBank (Accession No. KX196317).
Pairwise sequences analysis indicated that Pv-C7 had 68%, 55%, and 47% amino acid identity with the C7 proteins of C. idella (AEQ53931.1), Oplegnathus fasciatus (AFZ93893.1), and Sus scrofa (NP_999447.1), respectively. Multiple alignments showed that the Pv-C7 contained principally functional domains of the other C7 amino acid sequences, including two complement control protein modules (CCP; amino acids 555-609, 613-665), a low density lipoprotein receptor class A domain (LDLa; 83-115), one membrane-attack complex/ perforin domain (MACPF; 216-432), two thrombospondin type 1 repeats (TSP1; 26-76, 487- 533), and two factor I membrane attack complex (FIMAC; 755-816, 682-742) domains (Fig. 2).
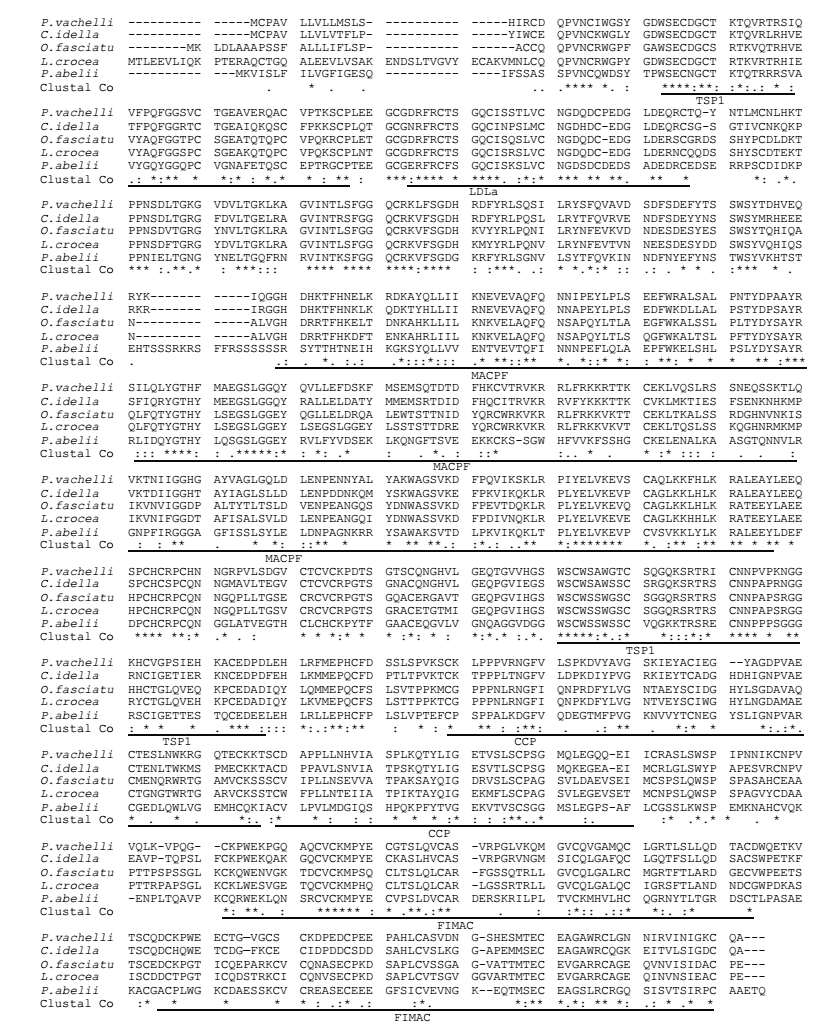
|
| Fig.2 Multiple alignments of the C7 protein The amino acid sequence from Pelteobagrus vachellii (Pv-C7) was aligned with that from Ctenopharyngodon idella (AEQ53931.1, 68%), Oplegnathus fasciatus (AFZ93893.1, 55%), Larimichthys crocea (KKF28532.1, 55%) and Pongo abelii (NP_001125756.1, 45%) using the Clustal W program. |
The mRNA expression of Pv-C7 in different tissues was detected in P. vachellii using the q-PCR assay, as shown in Fig. 3. The highest expression levels of PvC7 mRNA were observed in the liver tissue. The PvC7 mRNA was also detected in spleen, intestine, head kidney, gill and skin of P. vachellii, but not in other tissues analyzed.
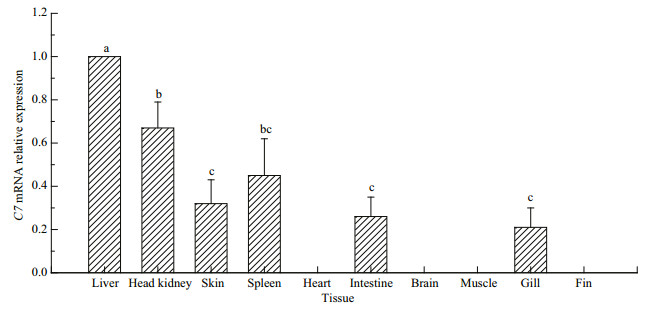
|
| Fig.3 Tissue expression profiles of Pv-C7 mRNA in Pelteobagrus vachellii Data are presented as the mean±SEM (n=9). Means with different superscripts represent statistically significant differences (P < 0.05). Differences in gene expression were analyzed by one-way analysis of variance (ANOVA) and multiple comparisons (Duncan) tests using SPSS software (SPSS v. 18.0). |
The RT-qPCR analysis showed that altering feeding time from 08:00 to 20:00 significantly increased the expression of CLOCK mRNA in liver and intestine (P < 0.05). It also increased the expression of CLOCK mRNA in spleen and head kidney, not appearing significant difference (P > 0.05) (Fig. 4a). Moreover, when shifting feeding to nighttime, the Pv-C7 mRNA significantly increased in the liver, spleen, intestine and head kidney tissues (P < 0.05) (Fig. 4b).
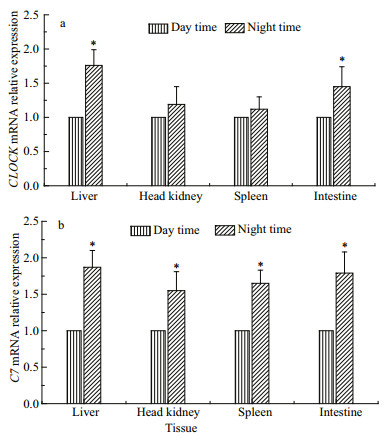
|
| Fig.4 Expression profiles of CLOCK (a) and Pv-C7 (b) mRNA in Pelteobagrus vachellii fed either only at day time or at night time Data are presented as the mean±SEM (n=12). Means with asterisk superscripts are significantly different between day time and night time (P < 0.05). The results of mRNA expression were analyzed by independent-sample t-tests (SPSS v. 18.0). |
In the two bacterial challenge groups, Pv-C7 mRNA in liver was significantly up-regulated from 48 to 96 h after A. hydrophila challenge (P < 0.05) (Fig. 5a), and C7 mRNA in head kidney and spleen tissues was found at obviously higher levels from 12- to 96-h post challenge, with highest expression levels at 48 h (P < 0.05) (Fig. 5b, c). Moreover, with the bacterial challenge, Pv-C7 mRNA expression from 48 to 96 h in the liver tissues of fish fed in the daytime appeared lower than that fed in the nighttime, although the differences were not significant (P > 0.05) (Fig. 5a). However, Pv-C7 mRNA expression from 12 to 48 h in head kidney tissues of fish fed in the daytime was significantly lower than that fed in the nighttime (P < 0.05) (Fig. 5b), and the same result appeared in spleen tissues from 12 to 24 h (P < 0.05) (Fig. 5c).
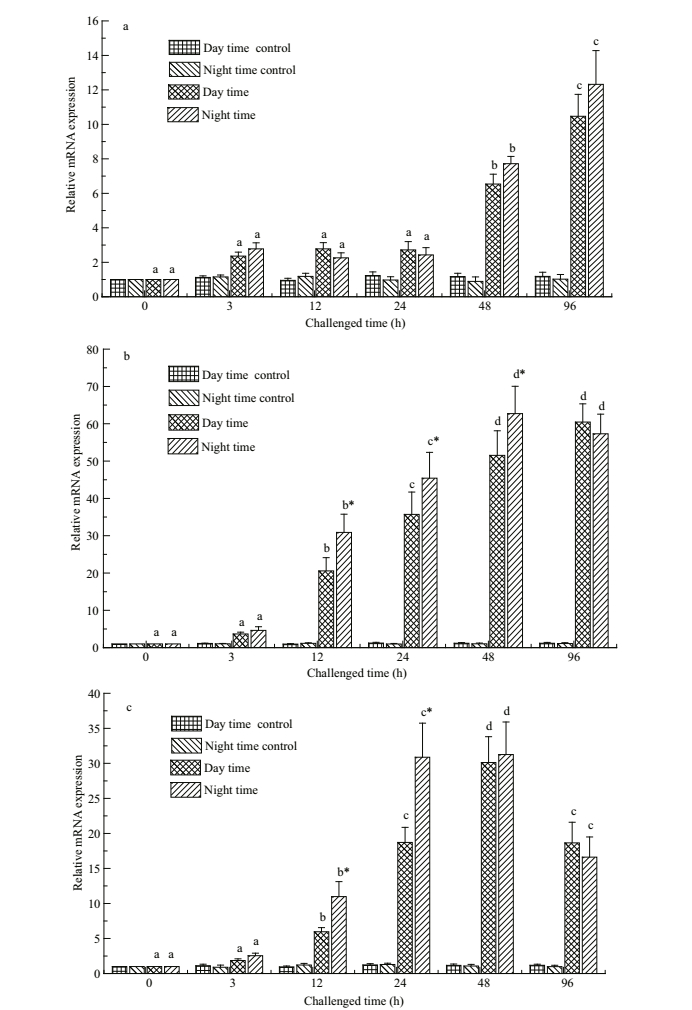
|
| Fig.5 Expression profiles of Pv-C7 mRNA in liver (a), head kidney (b) and spleen (c) tissue of Pelteobagrus vachellii fed either only at daytime or at nighttime, after challenge with A. hydrophila Data are presented as the mean±SEM (n=9). Means within the same treatment with different superscripts represent statistically significant differences (P < 0.05). Means with asterisk superscripts are significantly different between day time and night time within the same time point (P < 0.05). |
The immune system is under direct circadian control by systemic cues and molecular clocks within immune cells (Arjona and Sarkar, 2006; Logan and Sarkar, 2012; Phillips et al., 2015). A number of studies have indicated that circadian misalignment, which resulted from environmental desynchronization, shifting work or shifting feed, led to metabolic syndrome and diseases (Knutsson, 2003; Gao et al., 2009; Logan and Sarkar, 2012). Although complement components have been considered to play an essential role in immune system, and the investigation of complement components in fish has highlighted in molecular structure and expression with infection (Guo et al., 2016; Wang et al., 2015; Shen et al., 2012), the knowledge of their molecular expression was very limited especially in fish with an alternative feeding time. Therefore, in this study, the full-length sequence of C7 was identified in P. vachellii, and C7 mRNA expressions were firstly analyzed subject to bacterial challenge with an alternative feeding time.
Similar to human C7 and C. idella C7 (González et al., 2003; Shen et al., 2012), the SMART domain analysis showed that the Pv-C7 mature protein mainly consisted of eight domains (Fig. 2). Following the multiple alignment, these structural motifs were also observed in all the detected C7 homologues of C. idella, M. miiuy, L. crocea, and O. Fasciatus, including the C7 deduced amino acid sequences (Shen et al., 2012; Niroshana Wickramaarachchi et al., 2013; Wang et al., 2015; Guo et al., 2016). The others motifs of Pv-C7 showed extensive identity to the corresponding region in the other C7s. The C7 molecule indicated high conservation and integral function across species in the assembly of the MAC on the surface of complement-opsonized cells (Bossi et al., 2009).
In accordance with results obtained from C. idella and O. fasciatus, the highest expression level of C7 mRNA was found in the liver tissue (Fig. 3). Previous reports found similarly high C7 expression levels in liver tissues of C. idella, M. miiuy and mammals (González et al., 2003). From these studies, it seemed almost certain that the liver was the main production site of C7 (Witzel-Schlomp et al., 2001). The results suggested that Pv-C7 may act similarly to the other complement components that were generally of hepatic origin. The Pv-C7 mRNA was also detected in the spleen, intestine, head kidney, skin and gill of P. vachellii, but not in heart, brain and fin tissues. Similarly, C7 expression was found in all detected tissues except the heart in L. crocea, and it was ubiquitously expressed in all ten tested tissues in M. miiuy and O. fasciatus, although the levels of expression among the tissues were variable (Niroshana Wickramaarachchi et al., 2013; Wang et al., 2015; Guo et al., 2016). There was no C7 expression in the blood, brain, muscle, gill or fin tissues of C. idella (Shen et al., 2012). The different expression pattern in tissue resulted from specificity in different species, which need further investigation.
Del Pozo et al. (2012) reported that feeding rhythm (diurnal vs. nocturnal) strongly influenced CLOCK gene mRNA expression in the liver (feeding-entrained clock) of European seabass Dicentrarchus labrax (Del Pozo et al., 2012). Similarly, with a shift of feeding time to night, the CLOCK mRNA showed significantly increase in the liver and intestine of P. vachellii (P < 0.05). This increase also appeared in spleen and head kidney, without significant difference (P > 0.05) (Fig. 4a). This result indicated an altering feeding time could impact clock rhythms of peripheral tissues in P. vachellii. In addition, the CLOCK genes may also modulate the response of innate and adaptive immune systems (Logan and Sarkar, 2012), by regulating the level of cytokines and other factors, through direct control of the NF-κB signaling pathways (Scheidereit, 2006; Bozek et al., 2009). Interestingly, the putative transcription factor binding sites for NF-κB were found in the promoters of other TCC genes in C. idella (C9 and C7) (Li et al., 2007; Niroshana Wickramaarachchi et al., 2013), which suggested that the CLOCK gene could regulate TCC genes by NF-κB, including C7. This study proved that the significant increase of the expression of C7 mRNA appeared in the liver, spleen, intestine and head kidney tissues when altering feeding time from 08:00 to 20:00 (Fig. 4b). This result indicated that feeding time did affect the Pv-C7 mRNA expression. Similarly, the serum alkaline phosphatase (ALP), lysozyme (LYZ), peroxidase (PER) and protease (PRO) exhibited significant rhythmicity under 12L/12D cycles in Nile tilapia (Oreochromis niloticus) (Lazado et al., 2016). Additionally, in serum of shifted mice with treatment of four consecutive weekly 6-h phase advances of the light/dark schedule, several key immune factors were up-regulated, including IL-6, macrophage inflammatory protein- 2 (MIP-2), and leukemia inhibitory factor (LIF) (Castanon-Cervantes et al., 2010). Although the exact interactions and functions were still unknown, Arjona and Sakar (2006) hypothesized that, to promote the efficiency of the immune response, the coordination of immune factors was under the direct regulation of the molecular clock (Arjona and Sarkar, 2006). These studies indicated that altering the feeding time firstly altered the circadian expression of CLOCK in peripheral tissues, and then altered the circadian rhythm of the C7 expression, which was regulated by the CLOCK gene; but this hypothesis needed further investigation.
After challenging with A. hydrophila, an upregulation of mRNA was observed in the liver, spleen and head kidney tissue, in Pv-C7 comparison to the control group (Fig. 5). Similarly, Wang et al. (2015) indicated that expression levels of C7-1 in miiuy croaker were higher after exposing to Vibrio anguillarum and LPS (Wang et al., 2015). Significant changes in C7 transcript expression (> 20-fold) were observed following A. hydrophila infection in C. idella (Shen et al., 2012). Rapid and drastic responses to Vibrio alginolyticus challenges were observed for C7 in L. crocea (Guo et al., 2016). Moreover, perturbations of circadian rhythms by either external stressors, or internal stressors (e.g., psychological perturbations), may negatively affect health by harming immune function (Logan and Sarkar, 2012). For example, interleukin-17 (IL-17) in plasma was significantly increased than that of controls (Phillips et al., 2015). In serum of shifted mice with treatment of four consecutive weekly 6-h phase advances of the light/dark schedule, IL-6, leukemia inhibitory factor (LIF) showed upregulation, and the circadian desynchronization resulted in uncoordinated inflammatory responses to LPS challenge, which led to increased mortality (Castanon-Cervantes et al., 2010). In present study, after A. hydrophila challenge, Pv-C7 mRNA expression of fish fed at daytime was lower than that at nighttime in spleen and head kidney tissues from 12 to 48 h (P < 0.05) (Fig. 5b, c), and in liver from 48 to 96 h (P > 0.05) (Fig. 5a). These differences of C7 mRNA expression in spleen and head kidney may result from the perturbations of clock rhythms of immune cell by shifting feeding time. The light-dark and feeding cycles are the most important factors that entrain biological rhythms in fish species (Montoya et al., 2010b). Feeding time could coordinate the biological rhythms in peripheral tissues, such as in the liver (del Pozo et al., 2012). Rhythms cues were transmitted to immune tissues by neural and endocrine signals, and then mediated immune responses (Logan and Sarkar, 2012). Therefore, if feeding time synchronized with daily rhythms of behavior and physiology in animals, it may increase the ability to resist pathogens. However, this hypothesis needs deep investigation. Similarly, Liu et al. (2006) found period 2 mutant mice can be relatively resistant to LPS-induced endotoxic shock, in part due to decrease in the levels of the proinflammatory cytokines, IFN-γ and IL-1β. However, at 96 h, there was no significant difference in C7 mRNA expression among the liver, spleen and head kidney tissues of the fish in this study (P > 0.05) (Fig. 5); maybe the circadian rhythm of the fish was mainly controlled by the light-entrained clock in the peripheral tissues, not the feedingentrained clock (Del Pozo et al., 2012), and light signal promoted phase coherence of peripheral clocks in the immune system, and also governed daily variations in immune function (Logan and Sarkar, 2012).
In conclusion, Pv-C7 was successfully cloned in P. vachellii. Pv-C7 gene expression might be upregulated with feeding at nighttime in four tissues. Pv-C7 mRNA expression of the fish fed at nighttime showed significantly higher than that at daytime in head kidney and in spleen with Aeromonas hydrophila infection. This study contributed to give proper consideration for further accurate feeding schedules in this species.
Arjona A, Sarkar D K. 2006. Evidence supporting a circadian control of natural killer cell function. Brain, Behavior, and Immunity, 20(5): 469-476.
DOI:10.1016/j.bbi.2005.10.002 |
Bossi F, Rizzi L, Bulla R, Debeus A, Tripodo C, Picotti P, Betto E, Macor P, Pucillo C, Würzner R, Tedesco F. 2009. C7 is expressed on endothelial cells as a trap for the assembling terminal complement complex and may exert antiinflammatory function. Blood, 113(15): 3 640-3 648.
DOI:10.1182/blood-2008-03-146472 |
Bozek K, Relógio A, Kielbasa S M, Heine M, Dame C, Kramer A, Herzel H. 2009. Regulation of clock-controlled genes in mammals. PLoS One, 4(3): e4882.
DOI:10.1371/journal.pone.0004882 |
Castanon-Cervantes O, Wu M W, Ehlen J C, Paul K, Gamble K L, Johnson R L, Besing R C, Menaker M, Gewirtz A T, Davidson A J. 2010. Dysregulation of inflammatory responses by chronic circadian disruption. The Journal of Immunology, 185(10): 5 796-5 805.
DOI:10.4049/jimmunol.1001026 |
del Pozo A, Montoya A, Vera L M, Sánchez-Vázquez F J. 2012. Daily rhythms of clock gene expression, glycaemia and digestive physiology in diurnal/nocturnal European seabass. Physiology & Behavior, 106(4): 446-450.
|
Fisheries and Fisheries Administration Bureau of the Ministry of Agriculture. 2017. China Fishery Statistical Yearbook. China Agriculture Press, Beijing. p.25.
|
Fujito N T, Sugimoto S, Nonaka M. 2010. Evolution of thioester-containing proteins revealed by cloning and characterization of their genes from a cnidarian sea anemone, Haliplanella lineate. Developmental & Comparative Immunology, 34(7): 775-784.
|
Gao B, Radaeva S, Park O. 2009. Liver natural killer and natural killer T cells: immunobiology and emerging roles in liver diseases. Journal of Leukocyte Biology, 86(3): 513-528.
DOI:10.1189/JLB.0309135 |
González S, Martínez-Borra J, López-Larrea C. 2003. Cloning and characterization of human complement component C7 promoter. Genes and Immunity, 4(1): 54-59.
DOI:10.1038/sj.gene.6363902 |
Guo B Y, Wu C W, Lv Z M, Liu C L. 2016. Characterisation and expression analysis of two terminal complement components: C7 and C9 from large yellow croaker, Larimichthys crocea. Fish & Shellfish Immunology, 51: 211-219.
|
Kalsbeek A, Scheer F A, Perreau-Lenz S, La Fleur S E, Yi C X, Fliers E, Buijs R M. 2011. Circadian disruption and SCN control of energy metabolism. FEBS Letters, 585(10): 1 412-1 426.
DOI:10.1016/j.febslet.2011.03.021 |
Katagiri T, Hirono I, Aoki T. 1999. Molecular analysis of complement component C8β and C9 cDNAs of Japanese flounder, Paralichthys olivaceus. Immunogenetics, 50(1-2): 43-48.
DOI:10.1007/s002510050684 |
Knutsson A. 2003. Health disorders of shift workers. Occupational Medicine, 53(2): 103-108.
DOI:10.1093/occmed/kqg048 |
Lazado C C, Skov P V, Pedersen P B. 2016. Innate immune defenses exhibit circadian rhythmicity and differential temporal sensitivity to a bacterial endotoxin in Nile tilapia (Oreochromis niloticus). Fish & Shellfish Immunology, 55: 613-622.
|
Li L, Chang M X, Nie P. 2007. Molecular cloning, promoter analysis and induced expression of the complement component C9 gene in the grass carp Ctenopharyngodon idella. Veterinary Immunology and Immunopathology, 118(3-4): 270-282.
DOI:10.1016/j.vetimm.2007.05.005 |
Li M F. 2010. Progress on study on biology of Pelteeobaagrus fulvidraco (Richardson). Modern Fisheries Information, 25(9): 16-22.
(in Chinese with English abstract) |
Liu J G, Malkani G, Shi X Y, Meyer M, Cunningham-Runddles S, Ma X J, Sun Z S. 2006. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infection and Immunity, 74(8): 4 750-4 756.
DOI:10.1128/IAI.00287-06 |
Liu W, Jiang L H, Dong X L, Liu X X, Kang L S, Wu C W. 2016. Molecular characterization and expression analysis of the large yellow croaker (Larimichthys crocea) complement component C6 after bacteria challenge. Aquaculture, 458: 107-112.
DOI:10.1016/j.aquaculture.2016.03.003 |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using Q real-time quantitative PCR and the 2-∆∆CT method. Methods, 25(4): 402-408.
DOI:10.1006/meth.2001.1262 |
Logan R W, Sarkar D K. 2012. Circadian nature of immune function. Molecular and Cellular Endocrinology, 349(1): 82-90.
DOI:10.1016/j.mce.2011.06.039 |
Montoya A, López-Olmeda J F, Garayzar A B S, SánchezVázquez F J. 2010a. Synchronization of daily rhythms of locomotor activity and plasma glucose, cortisol and thyroid hormones to feeding in Gilthead seabream (Sparus aurata) under a light-dark cycle. Physiology & Behavior, 101(1): 101-107.
|
Montoya A, López-Olmeda J F, Yúfera M, Sánchez-Muros M J, Sánchez-Vázquez F J. 2010b. Feeding time synchronises daily rhythms of behaviour and digestive physiology in gilthead seabream (Sparus aurata). Aquaculture, 306(1-4): 315-321.
DOI:10.1016/j.aquaculture.2010.06.023 |
Morgan B P, Marchbank K J, Longhi M P, Harris C L, Gallimore A M. 2005. Complement: central to innate immunity and bridging to adaptive responses. Immunology Letters, 97(2): 171-179.
DOI:10.1016/j.imlet.2004.11.010 |
Mukherji A, Kobiitaa A, Chambona P. 2015. Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proceedings of the National Academy of Sciences of the Unite States of America, 112(48): E6 683-E6 690.
DOI:10.1073/pnas.1519735112 |
Muller-Eberhard H J. 1986. The membrane attack complex of complement. Annual Review of Immunology, 4(1): 503-528.
DOI:10.1146/annurev.iy.04.040186.002443 |
Niroshana Wickramaarachchi W D, Whang I, Kim E, Lim B S, Jeong H-B, De Zoysa M, Oh M-J, Jung S-J, Yeo S-Y, Yeon K S, Park H-C, Lee J. 2013. Genomic characterization and transcriptional evidence for the involvement of complement component 7 in immune response of rock bream (Oplegnathus fasciatus). Developmental & Comparative Immunology, 41(1): 44-49.
|
Oishi K, Ohkura N, Kadota K, Kasamatsu M, Shibusawa K, Matsuda J, Machida K, Horie S, Ishida N. 2006. Clock mutation affects circadian regulation of circulating blood cells. Journal of Circadian Rhythms, 4: 13.
DOI:10.1186/1740-3391-4-13 |
Phillips D J, Savenkova M I, Karatsoreos I N. 2015. Environmental disruption of the circadian clock leads to altered sleep and immune responses in mouse. Brain, Behavior, and Immunity, 47: 14-23.
DOI:10.1016/j.bbi.2014.12.008 |
Qin C J, Gong Q, Wen Z Y, Yuan D Y, Shao T, Wang J, Li H T. 2017. Transcriptome analysis of the spleen of the darkbarbel catfish Pelteobagrus vachellii in response to Aeromonas hydrophila infection. Fish & Shellfish Immunology, 70: 498-506.
|
Qin C J, Shao T. 2015. The Clock gene clone and its circadian rhythms in Pelteobagrus vachelli. Chinese Journal of Oceanology and Limnology, 33(3): 597-603.
DOI:10.1007/s00343-015-4167-x |
Scheidereit C. 2006. IκB kinase complexes: gateways to NF- κB activation and transcription. Oncogene, 25(51): 6 685-6 705.
DOI:10.1038/sj.onc.1209934 |
Shen Y B, Zhang J B, Xu X Y, Fu J J, Li J L. 2012. Expression of complement component C7 and involvement in innate immune responses to bacteria in grass carp. Fish & Shellfish Immunology, 33(2): 448-454.
|
Sun Y Y, Wang R X, Xu T J. 2013. Conserved structural complement component C3 in miiuy croaker Miichthys miiuy and their involvement in pathogenic bacteria induced immunity. Fish & Shellfish Immunology, 35(1): 184-187.
|
Wang S C, Gao Y H, Shu C, Xu T J. 2015. Characterization and evolutionary analysis of duplicated C7 in miiuy croaker. Fish & Shellfish Immunology, 45(2): 672-679.
|
Wang S C, Wang R X, Xu T J. 2013. The evolutionary analysis on complement genes reveals that fishes C3 and C9 experience different evolutionary patterns. Fish & Shellfish Immunology, 35(6): 2 040-2 045.
|
Witzel-Schlomp K, Rittner C, Schneider P M. 2001. The human complement C9 gene: structural analysis of the 5' gene region and genetic polymorphism studies. European Journal of Immunogenetics, 28(5): 515-522.
DOI:10.1046/j.0960-7420.2001.00248.x |
Yang R B, Xie C X, Wei K J, Zheng W Y, Lei C S, Feng K. 2006. The daily feeding rhythms of juvenile yellow catfish, Pelteobagrus fulvidraco at different feeding frequencies. Journal of Huazhong Agricultural University, 25(3): 274-276.
(in Chinese with English abstract) |
Zheng K K, Zhu X M, Han D, Yang Y X, Lei W, Xie S Q. 2010. Effects of dietary lipid levels on growth, survival and lipid metabolism during early ontogeny of Pelteobagrus vachelli larvae. Aquaculture, 299(1-4): 121-127.
DOI:10.1016/j.aquaculture.2009.11.028 |
 2018, Vol. 36
2018, Vol. 36


