Institute of Oceanology, Chinese Academy of Sciences
Article Information
- NIU Sufang, ZHAI Yun, WU Renxie, ZHANG Haoran, TIAN Letian, DENG Jiaxin, XIAO Yao
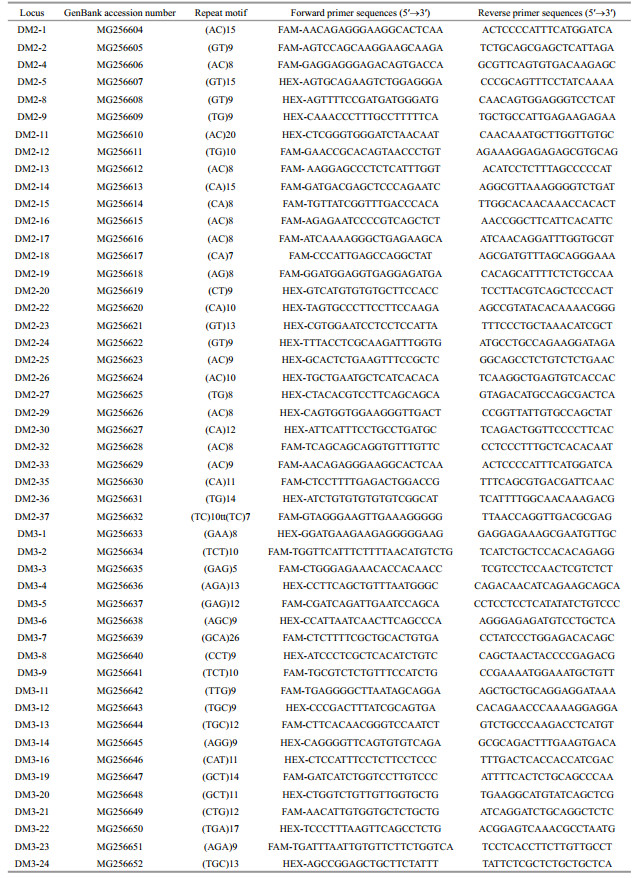
- Isolation and characterization of 49 polymorphic microsatellite loci for Decapterus maruadsi using SLAF-seq, and cross-amplification to related species
- Journal of Oceanology and Limnology, 37(1): 245-255
- http://dx.doi.org/10.1007/s00343-019-7299-6
Article History
- Received Oct. 28, 2017
- accepted in principle Dec. 6, 2017
- accepted for publication Jan. 9, 2019
The round scad, Decapterus maruadsi, is a warm-water pelagic fish of family Carangidae (Perciformes), which widely distributes in the coastal waters of west Pacific including China, Korea, and Japan (Liu et al., 2016). Recently, due to severe declines of previously favored high trophic level fishery resources, D. maruadsi has increasingly become one of the main commercial fishes (Chen et al., 2006; Zhang et al., 2007). Since the late 1990s, the annual catch of D. maruadsi in China has reached more than 500 000 tons, with a maximum of approximately 670 000 tons, ranking the third behind hairtail and anchovy (Ministry of agriculture fishery administration, 1999–2017). As a result, D. maruadsi has been heavily exploited in some areas. Consequently, of its biology characteristics have degraded, such as earlier sex maturation, smaller body length, and simpler population structure (Lu et al., 2000; Zhang et al., 2007). This has attracted our attention to genetic resources conservation of D. maruadsi. However, previous studies mainly focused on D. maruadsi fishery biology (Chen et al., 2006; Tong et al., 2012), and little is known about its population genetic and molecular markers except for mitochondrial gene (Li et al., 2016). In this regard, considerable attention has been paid to microsatellites, also known as simple sequence repeats (SSRs).
Microsatellite, an ideal single-locus co-dominant and hypervariable molecular marker, usually extensively distributed at random throughout the genome and has been widely applied in many research fields including population genetics, genome mapping, and parentage analyses (O'Connell and Wright, 1997; Stabile et al., 2016; Zhang et al., 2016). In the early days, microsatellite markers are developed by traditional methods such as magnetic bead enrichment (Khan et al., 2014), expressed sequence tag library (EST) (Henshaw et al., 2011), and genomic library (Chen et al., 2009), thus limiting the diversity and abundance of microsatellites obtained from the species genome (Restrepo et al., 2015). With the advent of next-generation sequencing (NGS), several reduced representation library (RRL) sequencing methods provide novel strategies for markers development (Altshuler et al., 2000; Shan et al., 2015). Among these, the recently developed specific-locus amplified fragment sequencing (SLAF-seq) enables the large-scale discovery of sequence-based marker in genome-wide and can strongly control marker number, frequency normalization, distribution, and repetitive regions in the absence of reference genome (Sun et al., 2013). In addition, SLAF-seq can create a balance between higher accuracy and relatively lower sequencing cost (Shan et al., 2015). Thus SLAF-seq technology is an efficient and high-resolution method for genome-wide microsatellite markers discovery, in particular in non-model species for which no reference genome information is available (Zhang et al., 2017a, b). To date, SLAF-seq has been successfully used to isolate microsatellites in several animals and plants, such as Lepturacanthus savala (Zhang et al., 2017a), Trichiurus japonicus (Zhang et al., 2017b), Boehmeria nivea (Luan et al., 2016), and Ormosia hosiei (Li et al., 2017). Accordingly, SLAF-seq technology can be generally applicable to develop a set of microsatellites for D. maruadsi.
In this study, the objective was to detect genomic microsatellites and isolate polymorphic microsatellite markers for D. maruadsi through SLAF-seq. Additionally, the isolated microsatellites were tested for their transferability in five phylogenetically related species from the family Carangidae. We expected the results could provide insights into further studies of population genetic, conservation genetics, and molecular phylogeny in D. maruadsi and other related species.
2 MATERIAL AND METHOD 2.1 Fish sample preparation and DNA extractionD. maruadsi (n=32, Shantou coast), Decapterus macarellus (n=1, Nansha Island), Decapterus kurroides (n=2, Sanya coast), Decapterus macrosoma (n=8, Sanya and Wenchang coast), Trachurus japonicus (n=5, Jiangmen coast), and Selaroides leptolepis (n=3, Beihai coast) were purchased from local fishermen in the South China Sea during the February 2010 and April 2015. After morphological identification, all specimens were preserved in 95% ethyl alcohol and kept at -20℃. Total genomic DNA was extracted from the muscle of each individual (Sambrook and Russell, 2002) for DNA sequencing, microsatellite screening, genotyping, and cross-species amplification. The quality, concentration, and purity of extracted DNA were monitored on 1% agarose gels.
2.2 SLAF library construction, SLAF-seq, and microsatellite searchThe SLAF-seq technology was used to construct library and sequence according to the procedures previously described by Sun et al. (2013) with minor modification as follows. An optimum enzyme digestion scheme was simulated with the genome of Lateolabrax japonicus as the reference. Pair-end sequencing was performed upon the selected SLAFs using an extracted DNA from D. maruadsi on Illumina HiSeqTM2500 (Illumina Inc., San Diego, CA, U.S.). The Oryza sativa indica was selected as a control to determine the reliability of this method. The potential microsatellite loci with motifs ranging from mono-nucleotide to hexa-nucleotide were identified and located using MIcroSAtellite identification tool (MISA) (http://pgrc.ipk-gatersleben.de/misa/). The minimum repeat unit was defined as follows: ten for mono-nucleotide, six for di-nucleotide, and five for all the higher order motifs (tri-, tetra-, penta-, and hexa-nucleotide). The loci with enough flanking sequence were used for primer design using Primer 3.0 software.
2.3 Primer polymorphism assessmentIn the study, we focused on the di-nucleotide and tri-nucleotide repeat loci due to their absolute predominance of quantity and high variability (Castoe et al., 2012). Finally, one hundred and thirty-two microsatellite markers (di- and tri-nucleotide) were randomly selected for the accuracy and polymorphism test in six randomly chosen D. maruadsi individuals. The polymerase chain reaction (PCR) was performed in a final volume of 15 μL containing 9.9 μL dH2O, 1.5 μL 10× EasyTaq® buffer for PAGE [200 mmol/Lol/L Tris-HCl (pH 8.3), 20 mmol/Lol/L MgSO4, 200 mmol/Lol/L KCl, 100 mmol/Lol/L (NH4)2SO4], 0.3 μL dNTP mixture (10 mmol/Lol/L), 0.3 μL forward primer (10 μmol/L), 0.3 μL reverse primer (10 μmol/L), 1.7 U EasyTaq® DNA polymerase for PAGE (TransGen Biotech Co. Ltd., Beijing, China), and 1 μL DNA (25–50 ng). PCR reactions were performed in a VeritiTM 96-Well Thermal Cycler (Applied Biosystems, USA) under the following parameters: initial denaturation at 95℃ for 4 min; followed by 25 cycles at 95℃ for 30 s, 62–52℃ for 30 s (decreasing 1℃ per cycle for the first 10 cycles), and 72℃ for 30 s, with a final extension at 72℃ for 10 min. The PCR reactions without the addition of the template DNA were used as blanks. The PCR products were separated by electrophoresis on 8% non-denaturing polyacrylamide gel and visualized by 0.1% silver nitrate staining. The pBR322 Marker/MspI marker (Tiangen, Beijing, China) was used for identifying allele size.
2.4 Microsatellite amplification and characterizationPolymorphic primers were amplified by three-primer PCR and further characterized using 32 D. maruadsi individuals collected from Shantou coast of Guangdong Province, the South China Sea during the spring of 2014. The 5′ end of each forward primer was labeled with an M13 tail (5′-AGGG-TTTTCCCAGTCACG-3′ or 5′-GAGCGGATAACA-ATTTCACAC-3′). FAM or HEX was added to the 5′ end of M13 universal primer which had the same sequence to the M13 tail. Three-primer PCR was determined as previously described (Wu et al., 2016), with slight modifications. Briefly, the PCR was amplified in 15 μL reaction volume with the following reagent volume: 9.8 μL dH2O, 1.5 μL 10 × EasyTaq® buffer for PAGE [200 mmol/L Tris-HCl (pH 8.3), 20 mmol/L MgSO4, 200 mmol/L KCl, 100 mmol/L (NH4)2SO4], 0.3 μL dNTP mixture (10 mmol/L), 0.3 μL forward primer (10 μmol/L) with M13 tail, 0.3 μL reverse primer (10 μmol/L), 0.1 μL labeled M13 universal primer (10 μmol/L), 1.7 U EasyTaq® DNA polymerase for PAGE (TransGen Biotech Co. Ltd., Beijing, China), and 1 μL DNA (25–50 ng). The following PCR conditions were used: an initial denaturation for 4 min at 95℃, followed by 25–28 cycles of 30 s at 95℃, 30s at locus-specific annealing temperature (52–62℃), 30 s at 72℃, and final elongation for 10 min at 72℃. All sets of PCR included a negative control reaction tube in which all reagents were included, except the template DNA. The PCR products were send to Shanghai Generay Biotech Company for genotyping using the Applied Biosystems 3730 DNA Analyzer and GeneMapper v 4.0.
2.5 Cross-species amplificationThe transferability of all developed microsatellite markers was checked on D. macarellus (n=1), D. kurroides (n=2), D. macrosoma (n=8), T. japonicus (n=5), and S. leptolepis (n=3) using the same amplification conditions as above. Fragment length analysis was carried out on 8% non-denaturing polyacrylamide gel, and allele size was identified by comparison with a pBR322 Marker/MspI marker (Tiangen, Beijing, China). The locus with at least one band of the expected size was considered to be transferable.
2.6 Data analysisThe number of alleles (Na), polymorphism information content (PIC), observed heterozygosity (Ho), and expected heterozygosity (He) were calculated with Cervus v 3.0.7 (Kalinowski et al., 2007). The Frequency of null alleles (Fua) was estimated by MicroChecker v 2.2.3 (Van Oosterhout et al., 2004) based on the Brookfield-1 method. FSTAT v 2.9.3 (Goudet, 2002) was used to evaluate inbreeding coefficient (Fis) of Weir and Cockerham's version. GenePop v 4.3 (Rousset, 2008) was used to test Hardy-Weinberg equilibrium (HWE) at each locus, and significance values (P) were adjusted for multiple comparisons with sequential Bonferroni correction (α=0.05) (Rice, 1989).
3 RESULT AND DISCUSSION 3.1 SLAF sequencing results and genomic microsatellites screeningUsing SLAF-seq technology, a total of 1.51 M pair-end sequence reads were generated in D. maruadsi. About 112 936 high-quality SLAFs were detected, with an average sequence depth of 6.21-fold. The sequencing quality score of 30 (Q30) and average guanine-cytosine (GC) content of genomic were about 82.32% and 44.97%, respectively, which indicated the relatively high quality and reliability of the SLAF-seq data. Total size of examined sequences was 58 501 440 bp. A total of 18 771 sequences (1.24%) contained microsatellite motif. MISA detected 28 905 microsatellites, of which di-nucleotide was the most frequent (13 590, 47.02%), followed by mono-nucleotide (8 138, 28.15%), tri-nucleotide (5 727, 19.81%), tetra-nucleotide (1 104, 3.82%), penta-nucleotide (234, 0.81%), and hexa-nucleotide (112, 0.39%). It was not surprising that di-nucleotide repeats were the most frequent motif type, which was consistent with many other marine fishes (Wu et al., 2016). In addition, we abnegated mono-nucleotide repeats because of the difficulty to distinguish genuine mono-nucleotide repeat from polyadenylation products.
The abundance of specific repeat motifs had large differences. Among di-nucleotide repeats, the highest frequency motif was AC (2084), followed by TG (2061), CA (1710), GT (1439), AG (518), GA (429), CT (384), and TC (375), while other motifs were comparatively scarce. Of the tri-nucleotide, the most frequent motif was GAG (256), followed by CAG (200), GCT (178), CTG (171), CCT (168), AGC (165), GGA (157), AGG (140), GCA (140), CTC (135), TGC (133), TCC (122), TGT (109), and AAC (103). Some tri-nucleotide motifs less than 100 had not been scheduled. Of the tetra-nucleotide, penta-nucleotide, and hexa-nucleotide, the abundance of specific repeat motifs was lower (< 23). The tandem repeat unit number of most microsatellites was not high. The repeats with 6–10 copies were the most common among the di-nucleotide, and repeats with 5–8 copies were the most common among tri-nucleotide, tetra-nucleotide, penta-nucleotide, and hexa-nucleotide. Our results suggested that SLAF-seq technology really was a reliability, rapid, cost-effective, and easy approach to identify massive numbers of various potential microsatellites in the genome.
3.2 Development and characterization of microsatellite markersThe results of polymorphism detection among six D. maruadsi individuals showed that 85 of 132 loci were polymorphic (polymorphism rate of 64.39%). The percentage of polymorphic loci was a little higher in present study (64.39%) than the mean for several previous studies (59%, Sousa-Santos et al., 2015). In order to save costs, a set of 70 fluorescently labeled primers were synthesized and amplified by three primers PCR (TP-PCR). However, by the optimization of PCR conditions (annealing temperatures and cycle number) and characterizations assessment using 32 D. maruadsi individuals, 21 polymorphic loci were deserted due to light gel bands, low amplification success rate (< 85%) or unreliable amplification. Finally, 49 loci were successfully amplified in at least 28 D. maruadsi individuals. These 49 microsatellite loci sequences showed to be polymorphic and were submitted to the NCBI (GenBank accession numbers: MG256604-MG256652, Table 1).
Characterizations of 49 polymorphic microsatellite markers were showed in Table 2. Allele sizes varied between 110 bp and 309 bp, indicating less change of allelic drop-out because of the sample degradation (Gill et al., 1996). The average number of alleles per locus was 13.633, ranging from 4 to 25. The Ho and He ranged from 0.233 to 1.000 and from 0.374 to 0.959, with mean values of 0.738 and 0.836, respectively. Based on the viewpoint proposed by Gill et al. (1996), a total of 34 loci (Ho > 0.7) could be considered to have discriminating power of > 0.9. Eleven loci were supposed to be moderately discernibility with a number of Ho ranging from 0.531 to 0.688. Compared with heterozygosity of other fishes, the Ho and He in D. maruadsi were lower than that for Seriola quinqueradiata (Ho=0.90, He=0.89) (Ohara et al., 2003) but higher than those for Seriola dumerili (Ho=0.676, He=0.705) (Renshaw et al., 2007) and Trachinotus carolinus (Ho=0.70, He=0.69) (Seyoum et al., 2007). Such high polymorphism of the 49 loci was also reflected in their PIC values (0.341–0.941, mean=0.806). According to the criterion defined by Botstein et al. (1980), all 49 loci were highly informative (PIC > 0.5) except DM3-1 (0.25 < PIC=0.341 < 0.5, reasonably informative). Taken together, these results demonstrated that most developed microsatellite loci exhibited moderate to comparatively high genetic diversity in D. maruadsi individuals, which indicated that the markers were highly polymorphic and well-resolved.

|
The null alleles frequencies of 49 loci were between -0.064 and 0.280, which could be categorized into three classes according to the criteria suggested by Chapuis and Estoup (2007). The first category contained 29 loci (51.18%), which had a low null allele frequency (Fua < 0.05, negligible). These loci with high informativeness (PIC > 0.5) and moderate to high discriminating power (Ho=0.563–1.000) were desirable for further genetic studies of D. maruadsi with the exception of DM3-1. The second category (18 loci, 36.73%) had a moderate frequency of null allele (0.05≤Fua < 0.2). This type of loci may reduce the population genetic diversity and lead to overestimation of both genetic distance and FST (Chapuis and Estoup, 2007). Thus, we should use these loci cautiously to conduct population genetics analyses. In particular, DM2-5, DM2-23, DM2-30, DM2-35, DM2-36, DM2-37, DM3-5, DM3-7, DM3- 12, and DM3-23 deviated significantly from HWE after Bonferroni correction (adjusted P=0.001), which was also indicated by homozygote excess (Table 2, Ho < He). The third category contained DM3-9 and DM3-13 with a large null allele frequency (Fua≥0.2). The two loci also showed significant deviation from expectations under HWE. The results suggested that DM3-9 and DM3-13 were problematic and were not suitable for further research.
Above knowable, similar to other fish microsatellites (Villanova et al., 2015; Wu et al., 2016), null allele was also a ubiquitous characteristic in microsatellite loci of D. maruadsi and might be relevant to HWE deviation of above 12 loci. Moreover, the high positive Fis values (0.152–0.382) were estimated for the 12 loci (Table 2), which was probably another evidence of homozygote excess (Heras et al., 2016). Lastly, D. maruadsi has been heavily exploited in some areas, leading to a significant degradation in many characteristics (Lu et al., 2000; Zhang et al., 2007). Therefore, population degradation or natural selection could not be excluded as an explanation of HWE deviation despite high level genetic diversity of the tested population.
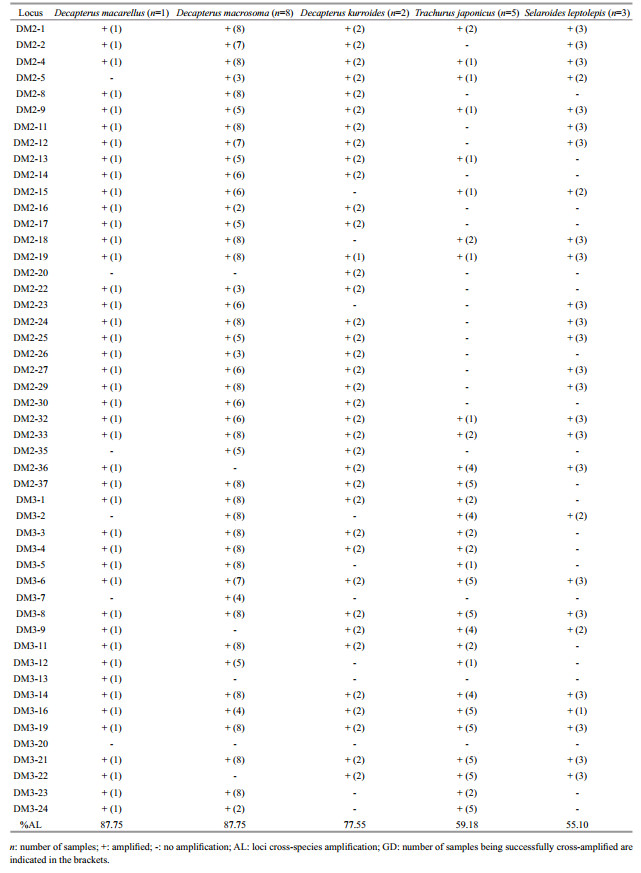
3.3 Cross-species transferabilityThe cross-species amplification results showed that 48 of 49 microsatellite loci successfully amplified across five species of the family Carangidae (Table 3), of which 12 successfully cross-amplified in five species, 19 in four species, 13 in three species, and 4 in one or two species. Thus, most of these loci were considered as highly versatile genetic markers and could provide new insights into further research of phylogenetic relationship among Carangidae species.
|
The cross-species amplification across three species of the genus Decapterus suggested a high transferability of 49 microsatellite markers from D. maruadsi to D. macarellus (43 loci, 87.76%), D. macrosoma (43 loci, 87.76%), and D. kurroides (38 loci, 77.55%). The loci transferability rates in genus Decapterus were slight lower than that in the Carangidae genus Seriola (100%, Babbucci et al., 2006) but significantly higher than those in the Sillaginidae genus Sillago (63.6%, Umino et al., 2013; 72.2%, Wu et al., 2016) and Trichiuridae genus Trichiurus (38.90%–60.71%, Zhang et al., 2017b). Furthermore, 32 loci successfully amplified across above three species, 12 across two species, and only 4 across one species, indicating a high conservation of flanking microsatellite regions in genus Decapterus. The present microsatellite loci provided the first known set of microsatellite DNA markers for D. macarellus, D. kurroides, and D. macrosoma.
Compared with genus Decapterus (77.55%–87.76%), relatively lower cross-amplification success within T. japonicus (29 loci, 59.18%) and S. leptolepis (27 loci, 55.10%) was achieved. Moreover, in order to examine the potential relationship between the cross- species amplification success and genetic distance, pairwise genetic distance between D. maruadsi and five confamilial species was estimated using MEGA v6.06 (Tamura et al., 2013) based on Cyt b (402 bp) and COI (651 bp) of D. maruadsi (KJ004518), D. macarellus (KM986880), D. kurroides (KJ464981 and JX26 1617), D. macrosoma (KF841444), T. japonicus (AP003092), and S. leptolepis (KU159666). The concatenated sequences (Cyt b + COI) genetic distance between D. maruadsi and S. leptolepis was 15.91%, which was higher than that between D. maruadsi and T. japonicus (10.81%), while the lowest were showed between D. maruadsi and three congeneric species D. macarellus, D. macrosoma and D. kurroides (8.90%, 9.10% and 10.06%, respectively). Obviously, the cross-species amplification success decreased with increasing genetic distance, demonstrating a negative relationship between genetic distances from D. maruadsi and cross-species microsatellite amplification success. Similar results have been reported for Serranus cabrilla (Carreras-Carbonell et al., 2008), Desmophyllum dianthus (Addamo et al., 2015), and Sillago japonica (Wu et al., 2016). In general, the transferability tests were successful in validating the utility of the markers within and between genera, indicating their effectiveness for further genetic studies. It is noteworthy negative correlations are also found between the percentage of polymorphic loci and the genetic divergence (Carreras-Carbonell et al., 2008). Consequently, the polymorphism of these loci having been successfully cross-amplified should be confirmed.
4 CONCLUSIONIn summary, this study confirmed that SLAF-seq technology was an effective method for microsatellite identification, and we had isolated the first set of novel microsatellite markers in D. maruadsi using this technology. The majority of them were highly polymorphic and well-resolved, suggesting an outstanding quality. Moreover, 48 of 49 loci developed here successfully cross-amplified in five other related species. These microsatellite loci would not only serve as valuable genetic tools for population genetic and molecular phylogeny, but also could play a crucial role in developing holistic conservation and fishery management strategies for the understudied species. It is notable that shorter repeat motifs (di- and tri-nucleotide) lead to possible misclassification of alleles (Castoe et al., 2012). In order to facilitate more accurate interpretation of allele lengths, longer repeat motif classes (tetra-, penta-, and hexa-nucleotide) should be developed in future studies.
5 DATA AVAILABILITY STATEMENTThe microsatellite sequences presented here are deposited in GenBank (accession numbers MG256604–MG256652).
Addamo A M, García-Jiménez R, Taviani M, Machordom A. 2015. Development of microsatellite markers in the deep-sea cup coral Desmophyllum dianthus by 454 sequencing and cross-species amplifications in Scleractinia order. Journal of Heredity, 106(3): 322-330.
DOI:10.1093/jhered/esv010 |
Altshuler D, Pollara V J, Cowles C R, Van Etten W J, Baldwin J, Linton L, Lander E S. 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature, 407(6803): 513-516.
DOI:10.1038/35035083 |
Babbucci M, Zane L, Andaloro F, Patarnello T. 2006. Isolation and characterization of microsatellite loci from yellowtail Seriola dumerilii (Perciformes:Carangidae). Molecular Ecology Notes, 6(4): 1 126-1 128.
DOI:10.1111/j.1471-8286.2006.01459.x |
Botstein D, White R L, Skolnick M, Davis R W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32(3): 314-331.
|
Carreras-Carbonell J, Macpherson E, Pascual M. 2008. Utility of pairwise mtDNA genetic distances for predicting cross-species microsatellite amplification and polymorphism success in fishes. Conservation Genetics, 9(1): 181-190.
DOI:10.1007/s10592-007-9322-2 |
Castoe T A, Streicher J W, Meik J M, Ingrasci M J, Poole A W, De Koning A P J, Campbell J A, Parkinson C L, Smith E N, Pollock D D. 2012. Thousands of microsatellite loci from the venomous coralsnake Micrurus fulvius and variability of select loci across populations and related species. Molecular Ecology Resources, 12(6): 1 105-1 113.
DOI:10.1111/1755-0998.12000 |
Chapuis M P, Estoup A. 2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution, 24(3): 621-631.
DOI:10.1093/molbev/msl191 |
Chen G B, Li Y Z, Zhao X Y, Chen Y Z, Jin X S. 2006. Acoustic assessment of five groups commercial fish in South China Sea. Acta Oceanologica Sinica, 28(2): 128-134.
(in Chinese with English abstract) |
Chen S L, Xing S C, Xu G B, Liao X L, Yang J F. 2009. Isolation and characterization of 10 polymorphic microsatellite loci from small yellow croaker(Pseudosciaena polyactis). Conservation Genetics, 10(5): 1 469-1 471.
DOI:10.1007/s10592-008-9762-3 |
Gill P, Urquhart A, Millican E, Oldroyd N, Watson S, Sparkes R, Kimpton C P. 1996. A new method of STR interpretation using inferential logic-development of a criminal intelligence database. International Journal of Legal Medicine, 109(1): 14-22.
DOI:10.1007/BF01369596 |
Goudet J. 2002. FSTAT: a program to estimate and test gene diversities and fixation indices (Version 2.9.3.2). https://www2.unil.ch/popgen/softwares/fstat.htm. Accessed on 2017-07-27.
|
Henshaw M T, Toth A L, Young T J. 2011. Development of new microsatellite loci for the genus Polistes from publicly available expressed sequence tag sequences. Insectes Sociaux, 58(4): 581-585.
DOI:10.1007/s00040-011-0164-z |
Heras S, Planella L, Caldarazzo I, Vera M, García-Marín J L, Roldán M I. 2016. Development and characterization of novel microsatellite markers by Next Generation Sequencing for the blue and red shrimp Aristeus antennatus. PeerJ, 4(3): e2200.
DOI:10.7717/peerj.2200 |
Kalinowski S T, Taper M L, Marshall T C. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology, 16(5): 1 099-1 106.
DOI:10.1111/j.1365-294X.2007.03089.x |
Khan G, Zhang F Q, Gao Q B, Jiao X J, Fu P C, Xing R, Zhang J H, Chen S L. 2014. Isolation of 16 microsatellite markers for Spiraea alpina and S. mongolica (Rosaceae) of the Qinghai-Tibet Plateau. Applications in Plant Sciences, 2(1): 1 300 059.
DOI:10.3732/apps.1300059 |
Li F Q, Xie Y J, Zhou Z C, Chen H P, Chu X L. 2017. The distribution types and characteristics of SSR loci based on SLAF-seq in Ormosia hosiei genome. Molecular Plant Breeding, 15(5): 1 774-1 781.
(in Chinese with English abstract) |
Li M, Chen Z Z, Chen T, Xiong D, Fan J T, Liang P W. 2016. Whole mitogenome of the Japanese scad Decapterus maruadsi (Perciformes:Carangidae). Mitochondrial DNA Part A:DNA Mapping, Sequencing, and Analysis, 27(1): 306-307.
DOI:10.3109/19401736.2014.892089 |
Liu J, Wu R X, Kang B, Ma L. 2016. Fishes of Beibu Gulf. Science Press, Beijing, China.
(in Chinese)
|
Lu Z B, Dai Q S, Yan Y M. 2000. An estimation of resources of chub mackerel, round scad and other pelagic fish stocks in the Taiwan Strait and the adjacent waters. Journal of Fishery Sciences of China, 7(1): 41-45.
(in Chinese with English abstract) |
Luan M B, Yang Z M, Zhu J J, Deng X, Liu C C, Wang X F, Xu Y, Sun Z M, Chen J H. 2016. Identification, evaluation, and application of the genomic-SSR loci in ramie. Acta Societatis Botanicorum Poloniae, 85(3): 3 510.
DOI:10.5586/asbp.3510 |
Michael O, Jonathan M W. 1997. Microsatellite DNA in fishes. Reviews in Fish Biology and Fisheries, 7(3): 331-363.
DOI:10.1023/A:1018443912945 |
Ministry of Agriculture Fishery Administration. 1999-2017.China Fishery Statistical Yearbook: 1999-2017. China Agricultural Press, Beijing, China. (in Chinese)
|
Ohara E, Nishimura T, Sakamoto T, Nagakura Y, Mushiake K, Okamoto N. 2003. Isolation and characterization of microsatellite loci from yellowtail Seriola quinqueradiata and cross-species amplification within the genus Seriola. Molecular Ecology Notes, 3(3): 390-391.
DOI:10.1046/j.1471-8286.2003.00460.x |
Renshaw M A, Patton J C, Rexroad Ⅲ C E, Gold J R. 2007. Isolation and characterization of dinucleotide microsatellites in greater amberjack, Seriola dumerili. Conservation Genetics, 8(4): 1 009-1 011.
DOI:10.1007/s10592-006-9221-y |
Restrepo A, Páez V P, Vásquez A, Daza J M. 2015. Rapid microsatellite marker development in the endangered neotropical freshwater turtle Podocnemis lewyana(Testudines:Podocnemididae) using 454 sequencing. Biochemical Systematics and Ecology, 59: 220-225.
DOI:10.1016/j.bse.2015.01.017 |
Rice W R. 1989. Analyzing tables of statistical tests. Evolution, 43(1): 223-225.
DOI:10.1111/j.1558-5646.1989.tb04220.x |
Rousset F. 2008. GENEPOP'007:a complete re-implementation of the GENEPOP software for Windows and Linux. Molecular Ecology Resources, 8(1): 103-106.
DOI:10.1111/j.1471-8286.2007.01931.x |
Sambrook J, Russell D W. 2002. Molecular Cloning: A Laboratory Manual. Huang P T, Trans. 3rd ed. Science Press, Beijing, China. (in Chinese)
|
Seyoum S, Denison S H, Tringali M D. 2007. Isolation and characterization of 13 polymorphic microsatellite loci for the Florida pompano, Trachinotus carolinus. Molecular Ecology Notes, 7(1): 141-143.
DOI:10.1111/j.1471-8286.2006.01556.x |
Shan T F, Pang S J, Li J, Li X, Su L. 2015. Construction of a high-density genetic map and mapping of a sex-linked locus for the brown alga Undaria pinnatifida(Phaeophyceae) based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Genomics, 16: 902.
DOI:10.1186/s12864-015-2184-y |
Sousa-Santos C, Fonseca P J, Amorim M C P. 2015. Development and characterization of novel microsatellite loci for Lusitanian toadfish, Halobatrachus didactylus. PeerJ, 3: e731.
DOI:10.7717/peerj.731 |
Stabile J, Lipus D, Maceda L, Maltz M, Roy N, Wirgin I. 2016. Microsatellite DNA analysis of spatial and temporal population structuring of Phragmites australis along the Hudson River Estuary. Biological Invasions, 18(9): 2 517-2 529.
DOI:10.1007/s10530-016-1157-7 |
Sun X W, Liu D Y, Zhang X F, Li W B, Liu H, Hong W G, Jiang C B, Guan N, Ma C X, Zeng H P, Xu C H, Song J, Huang L, Wang C M, Shi J J, Wang R, Zheng X H, Lu C Y, Wang X W, Zheng H K. 2013. SLAF-Seq:an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One, 8(3): e58700.
DOI:10.1371/journal.pone.0058700 |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2 725-2 729.
DOI:10.1093/molbev/mst197 |
Tong Y H, Mai R L, Chen J M, Li X M. 2012. Survey and analysis of landings on Hainan Island. South China Fisheries Science, 8(6): 85-91.
(in Chinese with English abstract) |
Umino T, Ueno K, Mihara T, Koike M, Watanabe M, Ahmad-Syazni K, Ishitani M, Ohara K. 2013. Isolation of eleven polymorphic microsatellite loci for the endangered Sillago parvisquamis and cross-species amplification with Sillago japonica. Conservation Genetics Resources, 5(3): 771-773.
DOI:10.1007/s12686-013-9904-x |
Van Oosterhout C, Hutchinson W F, Wills D P M, Shipley P. 2004. MICRO-CHECKER:software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4(3): 535-538.
DOI:10.1111/j.1471-8286.2004.00684.x |
Villanova G V, Vera M, Díaz J, Martinez P, Calcaterra N B, Arranz S E. 2015. Isolation and characterization of 20 polymorphic microsatellite loci in the migratory freshwater fish Leporinus obtusidens (Characiformes:Anostomidae) using 454 shotgun pyrosequencing. Journal of Fish Biology, 86(3): 1 209-1 217.
DOI:10.1111/jfb.12632 |
Wu R X, Zhang H R, Niu S F, Zhai Y, Liu X F. 2016. Development of polymorphic microsatellites for Sillago sihama based on next-generation sequencing and transferability to Sillago japonica. Genetics and Molecular Research, 15(4): gmr15049046.
DOI:10.4238/gmr15049046 |
Zhang H R, Liang Z B, Wu R X, Niu S F, Ke Z Y, Su S, Wei H, Wang Q, Sun B. 2017b. Development of microsatellite loci for Hairtail (Trichiurus japonicus) by using SLAF-seq technology and cross-species amplification test.Genomics and Applied Biology. http://kns.cnki.net/kcms/detail/45.1369.Q.20170705.1459.004.html. (in press) (in Chinese with English abstract)
|
Zhang H R, Liang Z B, Wu R X, Niu S F, Liang Y, Wang Q, Wei H, Xiao Y, Sun B. 2017a. Microsatellite loci isolation in the Savalai Hairtail (Lepturacanthus savala) based on SLAF-seq technology and transferability in the related species. Genomics and Applied Biology. http://kns.cnki.net/kcms/detail/45.1369.Q.20170926.1457.002.html. (in press) (in Chinese with English abstract)
|
Zhang H R, Niu S F, Wu R X, Zhai Y, Tian L T. 2016. Development and characterization of 26 polymorphic microsatellite markers in Lateolabrax maculatus and cross-species amplification for the phylogenetically related taxa. Biochemical Systematics and Ecology, 66: 326-330.
DOI:10.1016/j.bse.2016.05.008 |
Zhang Q H, Cheng J H, Xu H X, Shen X Q, Yu G P, Zheng Y J. 2007. The Fishery Resources and Sustainable Utilization of the East China Sea. Fudan University Press, Shanghai, China.
(in Chinese)
|
 2019, Vol. 37
2019, Vol. 37