Institute of Oceanology, Chinese Academy of Sciences
Article Information
- YE Lingtong, YAO Tuo, WU Lin, LU Jie, WANG Jiangyong
- Morphological and molecular diagnoses of Polydora brevipalpa Zachs, 1933 (Annelida: Spionidae) from the shellfish along the coast of China
- Journal of Oceanology and Limnology, 37(2): 713-723
- http://dx.doi.org/10.1007/s00343-019-7381-0
Article History
- Received Jan. 10, 2018
- accepted in principle Apr. 2, 2018
- accepted for publication Jun. 5, 2018
2 Shanghai Ocean University, Shanghai 201306, China
Polydorids are a speciose group comprising more than 140 described species within nine genera belonging to the Spionidae (Blake, 1996; Walker, 2011; Radashevsky, 2012). Polydorids are widely distributed globally and occur in a wide variety of habitats ranging from sandy and muddy sediments to calcareous substrates (Blake and Kudenov, 1978; Radashevsky, 1993; Blake, 1996; Sato-Okoshi, 1999; Simon, 2011; David and Williams, 2012). Polydorids are distinguished from the other spionids by an enlarged fifth chaetiger with large modified spines (Blake, 1996). Nowadays, identification of morphospecies mainly relies on character combinations or the inclusion of arithmetical differences among morphological characters (Blake, 1996; Walker, 2011; Radashevsky, 2012). The morphology of the modified spines and companion chaetae of chaetiger 5, the shape of the prostomium and pygidium, the extent of the caruncle, presence or absence of median antenna and occurrence of modified spines in posterior notopodia are considered to be diagnostic characteristics (Blake, 1996; Walker, 2011; Surugiu, 2012). Even some morphological properties of live worms such as the pigmentation pattern on palps and bodies and the colour of worm body and pygidium can be critical for species identification (Sato-Okoshi and Abe, 2013). However, this taxonomic criterion is extremely difficult for those species that possess highly similar morphological characters to one another. Furthermore, obtaining intact worms for species identification is often difficult because of the fragility of live worms, particularly the shell-boring species extracted from burrows by breaking the hard calcareous shells (Sato-Okoshi, 1999). Delimiting the degree of intraspecific and interspecific variation in morphological characters is difficult (Blake, 1996; Walker, 2011). This can easily cause confusion in species identification, giving rise to synonyms or homonyms of several polydorid species. Therefore, species identification by relying on morphological characters is typically insufficient for determination. Molecular methods, especially application of molecular markers such as nuclear 18S rRNA gene and mitochondrial cytochrome c oxidase subunit I (CO1) gene become increasingly important for assisting in the morphological identification of polydorids (Rice et al., 2008; Simon et al., 2009; Sato-Okoshi and Abe, 2012, 2013; Sato-Okoshi et al., 2013; Teramoto et al., 2013; Radashevsky and Pankova, 2013; Ye et al., 2015, 2017).
DNA barcoding is the derivation of short DNA sequences that enable species identification, recognition, and discovery in a particular domain of life (Hebert et al., 2003; Bucklin et al., 2011). CO1 is the most frequently used DNA barcod for metazoans and is considered to be a good marker for the identification of known species because of its marked divergence between genetic distance within metazoan species versus that among species (Erséus and Kvist, 2007; Bucklin et al., 2011). Although there are numerous studies on the application of the CO1 gene for species identification and phylogenetic structure of annelids (Bely and Wray, 2004; Erséus and Kvist, 2007; Blank and Bastrop, 2009; Luttikhuizen and Dekker, 2010; Nygren et al., 2010; Nygren and Pleijel, 2011; Norlinder et al., 2012; Pérez-Losada et al., 2015), only two reports examine polydorids (Rice et al., 2008; Ye et al., 2017). Finding high genetic distances among CO1 gene sequences from three separate locations in North America, Rice et al. (2008) clearly demonstrated that the nominal Polydora cornuta Bosc, 1802 contained at least three cryptic species. Using CO1 gene as a molecular marker, Ye et al. (2017) successfully validated unidentified larvae from the aquatic environment through the known CO1 sequences of polydorid adults. Synonyms or homonym may be very common, where identification is based on morphology alone. For example, P. convexa Blake and Woodwick, 1972 should be synonymous with Dipolydora bidentata (Zachs, 1933) (Radashevsky, 1993; Blake, 1996), and P. neocaea Williams and Radashevsky, 1999 might be a synonym of P. haswelli Blake and Kudenov, 1978 (Radashevsky et al., 2006; Read, 2010; Surugiu, 2012). However, by using CO1 gene sequences that provide clear evidence of variation ranges within species versus among species, the problems associated with using morphological characters alone may be solved (Hebert et al., 2003; Bucklin et al., 2011).
In this study, we provide a detailed redescription of P. brevipalpa Zachs, 1933, collected from mollusc shells, using morphological observations through light microscopy and scanning electron microscopy (SEM). The nuclear 18S rRNA gene and mitochondrial CO1 gene were used as support for morphological polydorid identification. The two gene sequences were compared to determine which gene is best for species identification. This study was the first to use the CO1 sequences of polydorids for the assessment of the delimitation of intraspecific and interspecific variation and to link the adult worms of P. brevipalpa to egg capsules from different hosts and locations.
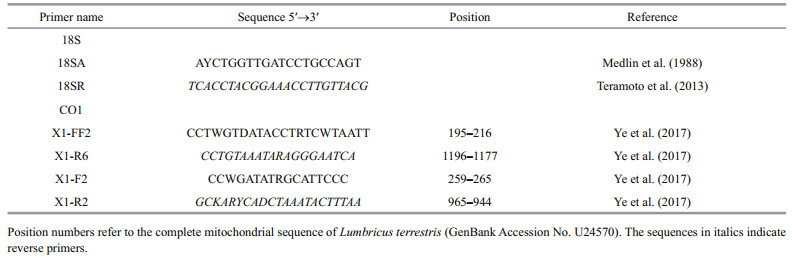
2 MATERIAL AND METHOD 2.1 Sampling and morphological observationsField collections were conducted from six sites along the coast of China (Fig. 1 and Table 1). Living bottom-sown scallops (Patinopecten yessoensis (Jay, 1857)) were randomly collected by scuba diving, and other mollusc shells, including wild and cultured Crassostrea gigas (Thunberg, 1793), were collected by hand from intertidal and shallow subtidal sites. All polydorid material was extracted by fracturing the calcareous mollusc shells with hammer and pliers, then transferred to the petri dish. Worms were relaxed by adding several drops of 5% MgCl2 solution and observed and photographed under a stereomicroscope SZX7 (Olympus, Japan) equipped with a digital camera. Egg capsules along the burrows of the worms were carefully collected using dissecting needles and forceps. Specimens were either fixed with 10% neutralised formalin for further morphological examination or fixed with 80% alcohol for molecular analysis. Specimens for SEM were fixed with 2.5% glutaraldehyde solution and dehydrated using a graded ethanol series, critical-point dried in carbon dioxide, coated with gold palladium, examined and then photographed using Hitachi S-3400N SEM.
|
| Fig.1 Map of the sampling sites along the coast of China Map drawing No. GS(2016)2891 (accessed from the National Administration of Surveying, Mapping and Geoinformation of China). |
|
One small fragment of each ethanol-preserved worm was cut for DNA extraction, and when necessary, whole egg capsules of the worms were used to obtain enough DNA. DNA was extracted using Tissue/Cell Genomic DNA Extraction Kit (BioTek, Beijing, China) according to the manufacturer's protocol.
Polymerase chain reaction (PCR) amplification of nuclear 18S rDNA and mitochondrial CO1 gene fragments was accomplished with the primers listed in Table 2. The primer pair X1-F2 and X1-R2 was used for some polydorid species where the primer pair X1-FF2 and X1-R6 did not work well. Each 50 μL reaction mixture contained 25 μL of 2× PCR mixture (Dongsheng, China), 2 μL of template, 19 μL of distilled water and 2 μL of each primer. Reaction mixtures were heated to 95℃ for 5 min, followed by 35 cycles of 50 s at 94℃, 50 s at 50℃ and 90 s for 72℃ and then a final extension of 10 min at 72℃ on Takara PCR Thermal Cycler Dice. The PCR products were sequenced in both directions using amplification primers and ABI Big Dye Terminator Chemistry on ABI 3730XL automatic DNA sequencer (Applied Biosystems, Inc., USA). Sequences of complementary strands were assembled using SeqMan 4.0 (DNAstar v7.0). The assembled sequences were checked and manually edited to avoid sequencing errors. The sequences were submitted to NCBI and deposited in GenBank (accession Nos. KF56224112, KP231289- 96, KP231302-5, KP231319-27, KP231331-6, KR052120-41).
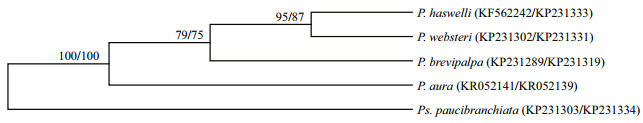
The 18S rDNA sequences of five polydorid species and their corresponding mitochondrial CO1 sequences obtained from GenBank (Table 3) were selected to compose 18S rDNA and mitochondrial CO1 data sets to calculate pairwise distances. Both data sets were aligned using Clustal X 1.83 (Thompson et al., 1997) with default parameters, and the resulting alignments were manually edited using the program BioEdit (Hall, 1999). Pairwise distances for intraspecific and interspecific polydorid species were calculated using MEGA 6.06 (Tamura et al., 2013) under default settings. One 18S rDNA sequence and one corresponding mitochondrial CO1 sequence of each polydorid species were selected to compose 18S rDNA tree-constructed data set and mitochondrial CO1 tree-constructed data set for construction of phylogenetic trees. After the alignments using BioEdit, the neighbour-joining phylogenetic trees based on 18S rDNA data and mitochondrial CO1 data were constructed in MEGA 6.06 using the neighbourjoining algorithm with pair-wise gap deletion. Clade support for both analyses was assessed through bootstrapping of 1 000 replicates. The phylogenetic tree is presented in Fig. 4.
|
| Fig.4 Phylogenetic tree generated by neighbour-joining analyses of 18S rDNA and mitochondrial CO1 sequences of 5 polydorid species The front and back numbers at nodes indicate bootstrap values calculated from 18S rDNA and CO1 datasets, respectively. The front and back GenBank accession numbers in brackets indicate 18S rDNA and CO1 sequences, respectively. P: Polydora; Ps: Pseudopolydora. |
|
A total of 30 mitochondrial CO1 sequences of P. brevipalpa were selected to analyse its genetic diversity and population genetic structure, including 20 CO1 sequences of adult worms and 10 CO1 sequences of egg capsules (Tables 1 and 3). The CO1 sequences were aligned using Clustal X 1.83 (Thompson et al., 1997) with default parameters. Haplotype diversity (h) and nucleotide diversity (π) were calculated using DnaSP5.0 (Librado and Rozas, 2009). The variation among and within the P. brevipalpa groups (using adult and egg capsules) was determined using analysis of molecular variance (AMOVA) in Arlequin version 3.5 (Excoffier and Lischer, 2010). A haplotype network was constructed in Network 4.613 (http://www.fluxus-engineering.) using the median joining network approach (Bandelt et al., 1999) with maximum parsimony calculation (Polzin and Daneschmand, 2003).
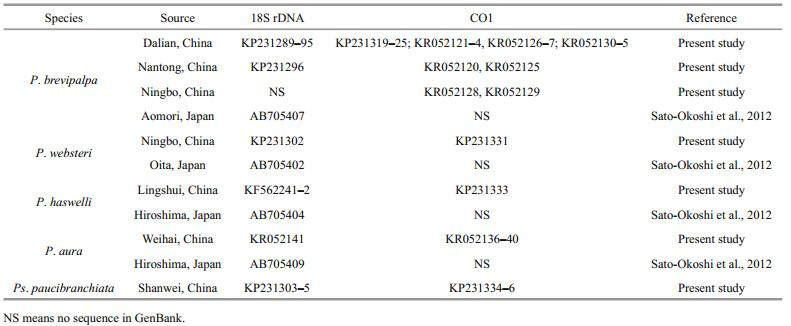
3 RESULT 3.1 Infestation condition of the host shellsHeavy infestation of P. brevipalpa was found in bottom-sown scallops Pa. yessoensis in the coastal waters of Zhangzi Island of Dalian (Fig. 1). Among the 50 randomly collected scallops, 90% were infested and 80% of the infested scallops were found to have more than 100 individuals in one host. On the outer surface of the infested shells, small holes were found across the entire valve. These holes were especially evident in the margin of the shell. Some sponges adhered to the outer surface of the valve. The inner surface of the valve was covered with a large number of U-shaped burrows (Fig. 2a). Some of these burrows were connected to one another, thereby forming some mud-blisters in the interlayer of the valve.
|
| Fig.2 Live worms of Polydora brevipalpa and infested shells of bottom-sown Patinopecten yessoensis a. inner surface of the valve, numerous burrows were observed inside the interlayer of the valve; Scale bar=10 mm; b. egg capsules, containing developing larvae, along the burrows of adult worms forming a long egg string; a single egg capsule indicated by the box; Scale bar=1 mm; c. live worms of P. brevipalpa, anterior end showing transverse black paired bands on the palps; Scale bar=1 mm; d. posterior chaetigers of P. brevipalpa showing disc-like pygidium; Scale bar=1 mm. |
Low prevalence and abundance of P. brevipalpa was observed in wild and cultured oysters (C. gigas) in the coastal waters of Nantong and Ningbo (Fig. 1 and Table 1). Of over 200 oysters collected from these two locations, only 5 were infested with P. brevipalpa, with P. websteri Hartman, 1943, P. haswelli and Boccardiella hamata (Webster, 1879) being the most common. A few P. brevipalpa were observed cooccurring with them. Species were difficult to identify because of the high diversity of polydorids and the difficulty in obtaining intact worms. In these two locations, a few adults and egg capsules of tentatively identified P. brevipalpa were collected and further identified using the molecular method (Table 1).
3.2 Morphological characterizationSystematics
Family Spionidae Grube, 1850
Genus Polydora Bosc, 1802
Polydora brevipalpa Zachs, 1933
Material examined
Zhangzi island, Dalian city, Liaoning Province, China (39°01′12.00″N, 122°45′0.00″E), from the shells of scallops (Patinopecten yessoensis (Jay, 1857)), coll. L. T. Ye, 24 Jun. 2014, NSB20150016, holytype; NSB20150017-20 (100+ spec.).
Description of new material
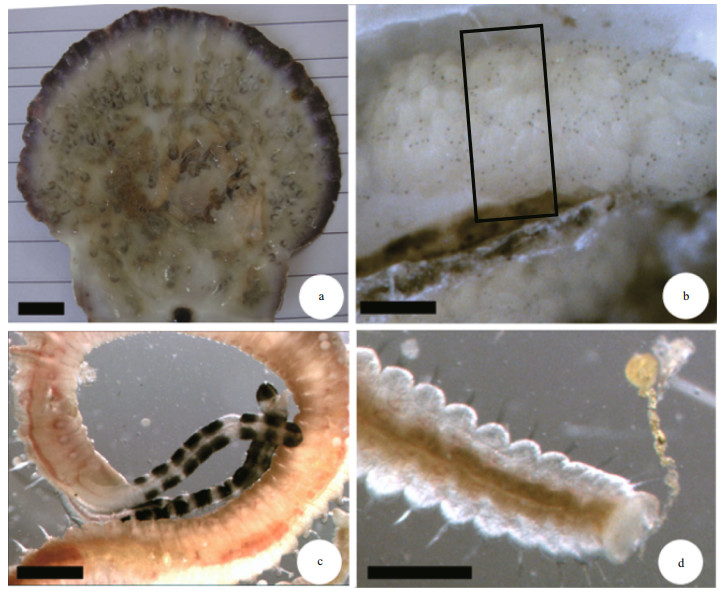
Measuring up to 20 mm long and 1.2 mm wide at chaetiger 5, with up to 110 chaetigers. Body light tan in life (Fig. 2c). Transverse black paired bands present, up to 8 on each palp (Fig. 2c). Spinulose papillae scattered inside the groove and on the margin of palps (Fig. 3a), visible with SEM only. Prostomium rounded anteriorly or weakly bilobed. Caruncle extending to the middle of chaetiger 3. One ciliary band row present on sides of caruncle (Fig. 3b). Four eyes present, trapezoidal in arrangement.
|
| Fig.3 Scanning electron microscopy images of Polydora brevipalpa a. palps showing spinulose papillae (arrowhead) inside the groove and on the margin; b. dorsal view of anterior end, caruncle extending to the middle of chaetiger 3 with each side of caruncle having one row of ciliary band (arrowhead); c. dorsal view of chaetiger 5 showing major modified spines with lateral flanges (arrowhead); d. dorsal view of chaetigers 9–13 showing branchiae with numerous cilia on the inner surface and a row of nototrochs (arrowhead) between a pair of branchiae on each chaetiger; e. lateral view of hooded hooks; f. lateral view of the posterior end showing disc-like pygidium. Scale bar=20 μm. |
Chaetiger 1 with capillary neurochaetae, notochaetae absent. Special spines on posterior chaetigers absent. Hooded hooks bidentate (Fig. 3e), with constriction on shaft, beginning from chaetiger 7, up to 10 per series, decreasing to 3 on posterior chaetigers (Fig. 3f). Branchiae from chaetiger 7, absent from the last several posterior chaetigers. Cilia scattered on the inner surface of branchiae. Nototrochs from chaetiger 7, each formed of one row of cilia located between pairs of branchiae on each chaetiger (Fig. 3d). Pygidium disc-like, occasionally cup-shaped (Figs. 2d, 3f).
Chaetiger 5 modified and enlarged, heavy spines alternating with pennoned companion chaetae. Fascicle of winged neurochaetae present, notochaetae absent. Heavy spines arranged in a slightly oblique row, 5–10 in number, falcate, with evident lateral flange or sheath (Fig. 3c).
Egg capsules present inside the burrows in some adult worms. Egg capsules joining each other, thus forming a long egg string (Fig. 2b). All the eggs developing synchronously into similar larvae.
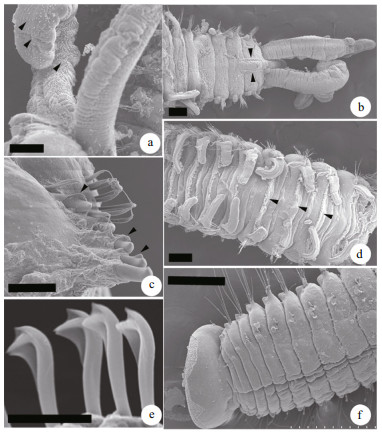
3.3 Molecular characterizationGood quality partial sequences of nuclear 18S rDNA and mitochondrial CO1 gene were obtained from 30 specimens of P. brevipalpa, including 20 adult worms and 5 egg capsules collected from Pa. yessoensis in Dalian and 3 and 2 egg capsules from C. gigas in Nantong and Ningbo, respectively (Table 1). Approximately 1 700 bp segment of 18S rDNA sequence and 648–924 bp segment of the CO1 gene sequence were successfully amplified. The 18S rDNA and the CO1 gene sequences of other polydorid species were either obtained using the same methods or directly acquired from GenBank to evaluate the divergence among polydorid species (Table 3). Genetic distance analyses showed that the intraspecific sequence divergence ranged from 0.2% to 0.9% based on the CO1 gene sequences and 0.0% to 0.4% based on 18S rDNA sequences (Table 4). The interspecific sequence divergence ranged from ranged from 16.7% to 25.6% based on the CO1 gene sequences and 0.5% to 5.6% based on 18S rDNA sequences (Table 4). The CO1 sequence divergence between polydorid species was evidently greater than that based on 18S rDNA sequences. The CO1 gene sequences of P. brevipalpa showed 99.4% to 100% sequence identity to one another and 79.3% (versus Pseudopolydora paucibranchiata (Okuda, 1937)) to 82.6% (versus P. websteri) sequence identity to other polydorid species. Phylogenetic analyses showed that the tree topology based on CO1 gene sequences was the same as that based on 18S rDNA sequences (Fig. 4).
|
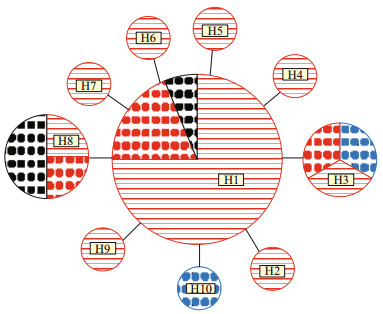
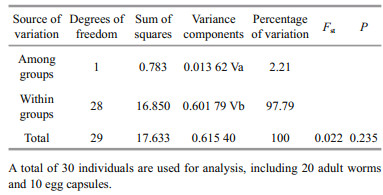
After editing all 30 CO1 sequences of P. brevipalpa, a 648-bp segment of sequences was retained for analysis. A total of 11 polymorphic sites were detected, and 10 haplotypes were identified. Hierarchical AMOVA test indicated that nearly all of the genetic variations (97.79%) were attributable to within groups of adult worms and egg capsules (Table 5). Only 2.21% of genetic variation was attributable to variability among groups of adult worms and egg capsules. No significant genetic structure (Fst=0.022, P=0.235) was detected among groups of adult worms and egg capsules (Table 5). A parsimony network showed that 7 out of 10 haplotypes were unique and represented by a single individual, and included 6 adult individuals from Dalian and 1 egg capsule from Ningbo (Fig. 5). The most common haplotype (H1) accounted for 53.3% (16 of 30) of all the individuals sampled, including 3 egg capsules and 12 adult worms from Pa. yessoensis in Dalian and 1 egg capsule from C. gigas in Nantong (Fig. 5). The haplotype H3 included one egg capsule and one adult worm from Pa. yessoensis in Dalian, as well as one egg capsule from C. gigas in Ningbo. The haplotype H8 shared four individuals, including one egg capsule and one adult worm from Pa. yessoensis in Dalian, as well as two egg capsules from C. gigas in Nantong (Fig. 5). The value of haplotype diversity (h) and nucleotide diversity (π) was 0.703±0.086 and 0.001 88±0.000 42, respectively. This result indicated that the CO1 sequences of P. brevipalpa had rather high levels of haplotype diversity and low nucleotide diversity.
|
| Fig.5 Haplotype network of Polydora brevipalpa based on mitochondrial CO1 sequences Circles represent haplotypes found, and their size is proportional to their frequency. Branch sizes are not proportional to genetic divergence. The area filled with red, black and blue represent the individuals collected from Dalian, Nantong and Ningbo, respectively. The individuals collected from adult worms and egg capsules are filled with transverse lines and dots, respectively. |
|
The results revealed that the interspecific variation (16.7%–25.6%) was at least 15 times larger than the intraspecific one (0.2%–0.9%) based on the CO1 gene sequences of polydorids (Table 4). However, 18S rDNA variation pattern demonstrated a rather narrow barcoding gap, with the interspecific variation (0.5%-5.6%) being very close to the intraspecific one (0.0%-0.4%) (Table 4). When comparing species within the genus Polydora based on 18S rDNA sequences, the barcoding gap even almost overlapped with the higher levels of intraspecific variation being close to the lower level of interspecific variation. The intraspecific variation of P. aura was 0.4%, whereas the interspecific variation between P. haswelli and P. websteri was only 0.5% (Table 4). Hebert et al. (2004) proposed a standard sequence threshold that the interspecific variation should be 10 times the mean intraspecific variation for the group understudy. In the present study, the CO1 gene exhibited better DNA barcode identification of polydorids than the 18S rDNA gene because of the sufficiently large barcoding gaps. In the CO1 gene sequence analyses of annelids, no universal criteria have currently equated to the differences relative to species boundaries. Erséus and Kvist (2007) assessed the CO1 variation in Scandinavian marine species of Tubificoides and demonstrated that the mean genetic distance within species was from 0.10% to 0.14% and between species was from 19.3% to 22.9%. Nygren et al. (2010) demonstrated that the intraspecific variation of Notophyllum crypticum Nygren et al., 2010 reached 3.3%, whereas the interspecific variation between N. crypticum and N. foliosum (Sars, 1835) was 8.5%. Nygren and Pleijel (2011) indicated that the genetic distances between Eumida sanguinea (Ørsted, 1843) species complex were in the range 6.5%–18.5%, and the intraspecific distances were in the range 0.2%-2.1%. After studying the taxonomy of the mud worm genus Marenzelleria based on the CO1 gene sequences, Blank and Bastrop (2009) found that the intraspecific variation within species of Marenzelleria was 0.2%–2.7%, whereas the interspecific variations were in the range 11.7%–21.7%. The present study showed a wide barcoding gap among polydorids based on the CO1 gene. However, the samples were from a limited geographic range in Chinese coastal waters, and the number of samples was also small. Addressing these two issues may result in narrowing the barcoding gaps among polydorids.
Polydora brevipalpa is considered to be a widely distributed polydorid species in the benthic coastal Asian waters of Japan, Russia and China (Radashevsky, 1993; Silina, 2006; Sato-Okoshi and Abe, 2013; SatoOkoshi et al., 2013). P. brevipalpa has been reported to be a specific borer of the scallop Pa. yessoensis (Radashevsky, 1993; Silina, 2006). However, Blake (1996) mentioned that P. brevipalpa was a shell borerinto various calcareous substrates in California, including bivalves and gastropod shells occupied by hermit crabs. Sato-Okoshi et al. (2013) also reported that P. brevipalpa can bore into the shells of abalone Haliotis discus hannai Ino, 1953 in Qingdao waters of China. In the present study, infestation by P. brevipalpa was observed in the shells of the oyster (C. gigas) in the waters of Ningbo and Nantong. Given the low infestation prevalence of P. brevipalpa on C. gigas, only a few incomplete individuals and egg capsules were collected for this study, hence the morphological identification at the species level was difficult. We compared the differences between P. brevipalpa in Ningbo and Nantong and Pa. yessoensis in Dalian based on the CO1 gene sequences to determine whether the former belongs or not to the same species with that collected from the shells of the latter. The AMOVA results based on CO1 gene sequences showed that most of the variation in sequences (97.79%) lay within groups of adult worms and egg capsules rather than between them. This result indicated that egg capsules from C. gigas in Ningbo and Nantong had a corresponding relationship to the adult worms from P. yessoensis in Dalian, and both belonged to P. brevipalpa. This result was further supported by parsimony network analysis, which showed that egg capsules collected from different localities and adult worms shared a single haplotype (Fig. 5). This study was the first to report P. brevipalpa infestation on C. gigas. As is commonly considered with other polydorids, this species was probably spread through transportation of infested Pa. yessoensis from Dalian to Nantong and Ningbo or effective transportation of planktonic larvae on ocean currents (Radashevsky and Olivares, 2005; Radashevsky et al., 2006; Sato-Okoshi et al., 2008; Simon et al., 2009).
Although adult polydorids have poor mobility, they have been frequently spread all over the world through commercial transportation or introduction of their host mollusc species (Radashevsky and Olivares, 2005; Simon et al., 2009; Sato-Okoshi et al., 2012). In the 1980s, the Japanese scallop Pa. yessoensis was introduced to China from Japan and is widely cultivated in the Northern waters of the Chinese coast (Li et al., 2007). P. brevipalpa (formerly known as P. variegata Imajima and Sato, 1984) was reported as the most dominant polydorid species boring into scallop Pa. yessoensis throughout the year in the Abashiri Bay of Japan (Mori et al., 1985; Sato-Okoshi et al., 1990). P brevipalpa (misidentified as P. ciliata (Johnston, 1838)) was recently reported as a kind of damaging pest, severely affecting the growth of Pa. yessoensis in Northern Chinese waters (Gao et al., 2011; Sato-Okoshi et al., 2013; Tang et al., 2015), and possibly brought to this region through transportation of Pa. yessoensis brood stock from Japan. Ascertaining whether P. brevipalpa is a native species in Chinese waters is difficult. P. brevipalpa appears to have the potential to successfully colonize this region and gradually spread to other mollusc species through human activity or transportation of planktonic larvae in the neighboring waters (Sato-Okoshi et al., 2013). Therefore, carefully checking for undesirable species is very important to avoid the introduction of harmful alien if introducing commercial brood stock.
So far, polydorid identification has been mainly based on the morphological characteristics of the adult worms (Walker, 2011). However, the morphological characteristics of most polydorids are variable, including prostomium shape, caruncle extent and pigmentation patterns of body and palps. For example, Teramoto et al. (2013) reported that P. onagawaensis Teramoto et al. (2013) showed highly variable pigmentation on the palps and body. Radashevsky and Nogueira (2003) demonstrated that the arrangement and shape of posterior notopodial spines of D. armata (Langerhans, 1880) was greatly variable, suggesting that D. armata is a junior synonym of P. rogeri (Martin, 1996). In the present study, we also observed variation in the shape of prostomium, the size-dependent extension of the caruncle and shape of pygidium in P. brevipalpa. The intraspecific variation of morphological characteristics made the species definition somewhat ambiguous, especially for those morphologically highly similar species. The application of nuclear 18S rDNA gene to confirm morphological taxonomy of polydorids was useful for identification and separation of those sibling species (Sato-Okoshi and Abe, 2012, 2013; Sato-Okoshi et al., 2013; Teramoto et al., 2013; Radashevsky and Pankova, 2013; Ye et al., 2015). However, the nuclear 18S rDNA gene is less useful to effectively identify sibling species. For example, the sequence identity between the sibling species P. calcarea (Templeton, 1836) (Accession No. AB705403) and P. onagawaensis (Accession No. AB69 1768) reached as high as 99.4% (1 761 nt/ 1 771 nt). The present study indicated that the CO1 gene had higher resolution than the nuclear 18S rDNA gene for polydorid identification. The wide barcoding gaps based on the CO1 gene sequences suggest that the CO1 gene is useful for polydorid identification and can be used to identify egg capsules, and possibly larvae.
Further studies are urgently required on the CO1 sequence characterization of polydorids from other geographical localities in different parts of the world. Sequencing CO1 from specimens collected in original type localities would also be useful to link the morphology to the molecular identity. Further research using the CO1 gene to link adult polydorids infesting molluscs to planktonic larvae in coastal sea waters may help to shine a light on the occurrence of this pest species globally.
5 CONCLUSIONIn this study, the CO1 gene was used for molecular identification of P. brevipalpa and the confirmation of the identity of polydorid egg capsules and the shellboring polydorid adult worms. Our results demonstrated that the CO1 gene is a useful DNA barcode for polydorid identification.
6 DATA AVAILABILITY STATEMENTThe datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
7 ACKNOWLEDGMENTWe are very grateful to Prof. JI Hongjiu (Institution of Oceanology and Marine Fishery, Jiangsu) and Prof. YAN Xiwu (Dalian Ocean University) for their assistance in the collection of samples. Special thanks are extended to two anonymous reviewers, who give us many constructive suggestions and critical comments, and spend a lot of energy to polish our manuscript.
Bandelt H J, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16(1): 37-48.
DOI:10.1093/oxfordjournals.molbev.a026036 |
Bely A E, Wray G A. 2004. Molecular phylogeny of naidid worms (Annelida:Clitellata) based on cytochrome oxidase I. Molecular Phylogenetics and Evolution, 30(1): 50-63.
DOI:10.1016/S1055-7903(03)00180-5 |
Blake J A, Kudenov J D. 1978. The Spionidae (Polychaeta) from southeastern Australia and adjacent areas with a revision of the genera. Memoirs of the National Museum of Victoria, 39: 171-280.
DOI:10.24199/j.mmv.1978.39.11 |
Blake J A. 1996. Family Spionidae Grube, 1850. Including a review of the genera and species from California and a revision of the genus Polydora Bosc, 1802. In: Blake J A, Hilbig B, Scott P H eds. Taxonomic Atlas of the Benthic Fauna of the Santa Maria Basin and Western Santa Barbara Channel. Volume 6. The Annelida Part 3-Polychaeta: Orbiniidae to Cossuridae. Santa Barbara Museum of Natural History, Santa Barbara, California.p.81-223.
|
Blank M, Bastrop R. 2009. Phylogeny of the mud worm genus Marenzelleria (Polychaeta, Spionidae) inferred from mitochondrial DNA sequences. Zoologica Scripta, 38(3): 313-321.
DOI:10.1111/zsc.2009.38.issue-3 |
Bucklin A, Steinke D, Blanco-Bercial L. 2011. DNA barcoding of marine metazoa. Annual Review of Marine Science, 3(1): 471-508.
DOI:10.1146/annurev-marine-120308-080950 |
David A A, Williams J D. 2012. Morphology and natural history of the cryptogenic sponge associate Polydora colonia Moore, 1907 (Polychaeta:Spionidae). Journal of Natural History, 46(23-24): 1 509-1 528.
DOI:10.1080/00222933.2012.679323 |
Erséus C, Kvist S. 2007. COI variation in Scandinavian marine species of Tubificoides (Annelida:Clitellata:Tubificidae). Journal of the Marine Biological Association of the United Kingdom, 87(5): 1 121-1 126.
|
Excoffier L, Lischer H E L. 2010. Arlequin Suite ver 3.5, a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10: 564-567.
DOI:10.1111/men.2010.10.issue-3 |
Gao Y, Zhang T, Yang H S, Zhang X F. 2011. The study on morphological and anatomic observation of Polydora ciliate. Marine Sciences, 35(10): 103-109.
(in Chinese with English abstract) |
Hall T A. 1999. BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95-98.
|
Hebert P D N, Cywinska A, Ball S L, deWaard J R. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B:Biological Sciences, 270(1512): 313-321.
DOI:10.1098/rspb.2002.2218 |
Hebert P D N, Stoeckle M Y, Zemlak S T, Francis C M. 2004. Identification of birds through DNA barcodes. PLoS Biology, 2(10): e312.
DOI:10.1371/journal.pbio.0020312 |
Li Q, Xu K F, Yu R H. 2007. Genetic variation in Chinese hatchery populations of the Japanese scallop (Patinopecten yessoensis) inferred from microsatellite data. Aquaculture, 269(1-4): 211-219.
DOI:10.1016/j.aquaculture.2007.04.017 |
Librado P, Rozas J. 2009. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1 451-1 452.
DOI:10.1093/bioinformatics/btp187 |
Luttikhuizen P C, Dekker R. 2010. Pseudo-cryptic species Arenicola defodiens and Arenicola marina (Polychaeta:Arenicolidae) in Wadden Sea, North Sea and Skagerrak:morphological and molecular variation. Journal of Sea Research, 63(1): 17-23.
DOI:10.1016/j.seares.2009.09.001 |
Medlin L, Elwood H J, Stickel S, Sogin M L. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene, 71(2): 491-499.
DOI:10.1016/0378-1119(88)90066-2 |
Mori K, Sato W, Nomura T, Imajima M. 1985. Infestation of the Japanese scallop Patinopecten yessoensis by the boring polychaetes, Polydora, on the Okhotsk Sea coast of Hokkaido, especially in Abashiri waters. Bulletin of Japanese Society of Science of Fishery, 51(3): 371-380.
DOI:10.2331/suisan.51.371 |
Norlinder E, Nygren A, Wiklund H, Pleijel F. 2012. Phylogeny of scale-worms (Aphroditiformia, Annelida), assessed from 18SrRNA, 28SrRNA, 16SrRNA, mitochondrial cytochrome c oxidase subunit I (COI), and morphology. Molecular Phylogenetics and Evolution, 65(2): 490-500.
DOI:10.1016/j.ympev.2012.07.002 |
Nygren A, Eklöf J, Pleijel F. 2010. Cryptic species of Notophyllum (Polychaeta:Phyllodocidae) in Scandinavian waters. Organisms Diversity and Evolution, 10(3): 193-204.
DOI:10.1007/s13127-010-0014-2 |
Nygren A, Pleijel F. 2011. From one to ten in a single stroke -resolving the European Eumida sanguinea (Phyllodocidae, Annelida) species complex. Molecular Phylogenetics and Evolution, 58(1): 132-141.
DOI:10.1016/j.ympev.2010.10.010 |
Pérez-Losada M, Breinholt J W, Aira M, Domínguez J. 2015. An updated multilocus phylogeny of the Lumbricidae(Annelida:Clitellata:Oligochaeta) earthworms. Journal of Phylogenetics and Evolutionary Biology, 3: 140.
|
Polzin T, Daneschmand S V. 2003. On Steiner trees and minimum spanning trees in hypergraphs. Operations Research Letters, 31(1): 12-20.
DOI:10.1016/S0167-6377(02)00185-2 |
Radashevsky V I, Lana P C, Nalesso R C. 2006. Morphology and biology of Polydora species (Polychaeta:Spionidae)boring into oyster shells in South America, with the description of a new species. Zootaxa, 1353: 1-37.
|
Radashevsky V I, Nogueira J M M. 2003. Life history, morphology and distribution of Dipolydora armata(Polychaeta:Spionidae). Journal of the Marine Biological Association of the United Kingdom, 83(2): 375-384.
DOI:10.1017/S0025315403007227h |
Radashevsky V I, Olivares C. 2005. Polydora uncinata(Polychaeta:Spionidae) in Chile:an accidental transportation across the Pacific. Biological Invasions, 7(3): 489-496.
|
Radashevsky V I, Pankova V V. 2013. Shell-boring versus tube-dwelling:is the mode of life fixed or flexible? Two cases in spionid polychaetes (Annelida, Spionidae). Marine Biology, 160(7): 1 619-1 624.
DOI:10.1007/s00227-013-2214-8 |
Radashevsky V I. 1993. Revision of the genus Polydora and related genera from the North West Pacific (Polychaeta:Spionidae). Publications of the Seto Marine Biological Laboratory, 36(1-2): 1-60.
DOI:10.5134/176224 |
Radashevsky V I. 2012. Spionidae (Annelida) from shallow waters around the British Islands:an identification guide for the NMBAQC Scheme with an overview of spionid morphology and biology. Zootaxa, 3152: 1-35.
DOI:10.11646/zootaxa.3152.1 |
Read G B. 2010. Comparison and history of Polydora websteri and P. haswelli (Polychaeta:Spionidae) as mud-blister worms in New Zealand shellfish. New Zealand Journal of Marine and Freshwater Research, 44(2): 83-100.
DOI:10.1080/00288330.2010.482969 |
Rice S A, Karl S, Rice K A. 2008. The Polydora cornuta complex (Annelida:Polychaeta) contains populations that are reproductively isolated and genetically distinct. Invertebrate Biology, 127(1): 45-64.
DOI:10.1111/j.1744-7410.2007.00104.x |
Sato-Okoshi W, Abe H. 2012. Morphological and molecular sequence analysis of the harmful shell boring species of Polydora (Polychaeta:Spionidae) from Japan and Australia. Aquaculture, 368-369: 40-47.
DOI:10.1016/j.aquaculture.2012.08.046 |
Sato-Okoshi W, Abe H. 2013. Morphology and molecular analysis of the 18S rRNA gene of oyster shell borers, Polydora species (Polychaeta:Spionidae), from Japan and Australia. Journal of the Marine Biological Association of the United Kingdom, 93(5): 1 279-1 286.
DOI:10.1017/S002531541200152X |
Sato-Okoshi W, Okoshi K, Abe H, Li J Y. 2013. Polydorid species (Polychaeta, Spionidae) associated with commercially important mollusk shells from eastern China. Aquaculture, 406-407: 153-159.
DOI:10.1016/j.aquaculture.2013.05.017 |
Sato-Okoshi W, Okoshi K, Koh B S, Kim Y H, Hong J S. 2012. Polydorid species (Polychaeta:Spionidae) associated with commercially important mollusk shells in Korean waters. Aquaculture, 350-353: 82-90.
DOI:10.1016/j.aquaculture.2012.04.013 |
Sato-Okoshi W, Okoshi K, Shaw J. 2008. Polydorid species(Polychaeta:Spionidae) in south-western Australian waters with special reference to Polydora uncinata and Boccardia knoxi. Journal of the Marine Biological Association of the United Kingdom, 88(3): 491-501.
|
Sato-Okoshi W, Sugawara Y, Nomura T. 1990. Reproduction of the boring polychaete Polydora variegata inhabiting scallops in Abashiri Bay, North Japan. Marine Biology, 104(1): 61-66.
|
Sato-Okoshi W. 1999. Polydorid species (Polychaeta:Spionidae) in Japan, with descriptions of morphology, ecology and burrow structure. 1. Boring species. Journal of the Marine Biological Association of the United Kingdom, 79(5): 831-848.
DOI:10.1017/S0025315498001003 |
Silina A V. 2006. Tumor-like formations on the shells of Japanese scallops Patinopecten yessoensis (Jay). Marine Biology, 148(4): 833-840.
DOI:10.1007/s00227-005-0120-4 |
Simon C A, Thornhill D J, Oyarzun F, Halanych K M. 2009. Genetic similarity between Boccardia proboscidea from Western North America and cultured abalone, Haliotis midae, in South Africa. Aquaculture, 294(1-2): 18-24.
DOI:10.1016/j.aquaculture.2009.05.022 |
Simon C A. 2011. Polydora and Dipolydora (Polychaeta:Spionidae) associated with molluscs on the south coast of South Africa, with descriptions of two new species. African Invertebrates, 52(1): 39-50.
DOI:10.5733/afin.052.0104 |
Surugiu V. 2012. Systematics and ecology of species of the Polydora-complex (Polychaeta:Spionidae) of the Black Sea. Zootaxa, 3518: 45-65.
|
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2 725-2 729.
DOI:10.1093/molbev/mst197 |
Tang B, Ye L T, Cao C, Yang B L, Wang J Y. 2015. Morphological and anatomic observation of Polydora brevipalpa in Patinopecten yessoensis. South China Fisheries Science, 11(4): 95-101.
(in Chinese with English abstract) |
Teramoto W, Sato-Okoshi W, Abe H, Nishitani G, Endo Y. 2013. Morphology, 18S rRNA gene sequence and life history of a new Polydora species (Polychaeta:Spionidae)from northeastern Japan. Aquatic Biology, 18(1): 31-45.
DOI:10.3354/ab00485 |
Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The CLUSTAL_X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24): 4 876-4 882.
DOI:10.1093/nar/25.24.4876 |
Walker L M. 2011. A review of the current status of the Polydora-complex (Polychaeta:Spionidae) in Australia and a checklist of recorded species. Zootaxa, 2751: 40-62.
DOI:10.11646/zootaxa.2751.1 |
Ye L T, Cao C, Tang B, Yao T, Wang R X, Wang J Y. 2017. Morphological and molecular characterization of Polydora websteri (Annelida:Spionidae), with remarks on relationship of adult worms and larvae using mitochondrial COI gene as a molecular marker. Pakistan Journal of Zoology, 49(2): 699-710.
DOI:10.17582/journal.pjz/2017.49.2.699.710 |
Ye L T, Tang B, Wu K C, Su Y L, Wang R X, Yu Z N, Wang J Y. 2015. Mudworm Polydora lingshuiensis sp. n is a new species that inhabits both shell burrows and mudtubes. Zootaxa, 3986: 88-100.
DOI:10.11646/zootaxa.3986.1 |
 2019, Vol. 37
2019, Vol. 37