Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Guodong, ZHANG Lili, XU Jianbo, YINC heng, ZHANG Ziping, WANG Yilei
- The roles of thyroid hormone receptor and T3 in metamorphosis of Haliotis diversicolor
- Journal of Oceanology and Limnology, 37(2): 745-758
- http://dx.doi.org/10.1007/s00343-019-7359-y
Article History
- Received Jan. 22, 2018
- accepted in principle Apr. 28, 2018
- accepted for publication Jun. 12, 2018
2 College of Animal Science, Fujian Agriculture and Forestry University, Fuzhou 350002, China
Extant metazoans have a wide variety of life cycles. In most marine invertebrates, hatching produces a larva. The larva continues to grow and develop to varying degrees. There appear dramatic morphological, physiological, behavioral and ecological transformations between the larvae and the juvenile (Bishop et al., 2006; Holstein and Laudet, 2014), which is defined metamorphosis. The juvenile grows and becomes an adult after experiencing sexual maturation.
Transitions between different states of development, physiology, and life history are typically mediated by hormones (Bishop et al., 2006; Flatt et al., 2006). Apart from insects and amphibians, our understanding of the molecular cascade controlling metamorphosis is very limited. In amphilbians and insects, the production of hormones (thyroid hormone in amphibians, ecdysone and juvenile hormone in insects) is controlled by the nerve system, as well as growth and environmental signals. These hormones control a complex tissue-specific gene regulatory network that drives metamorphosis (Holstein and Laudet, 2014).
Many studies have demonstrated that thyroid hormones (THs) regulate development, metamorphosis, and other life history transitions in marine invertebrates. For example, a TH derivative, TRIAC, induces metamorphosis in the Cephalochordate amphioxus (Paris et al., 2008). In echinoids, the TH thyroxine (T4) accelerates development to metamorphosis (Heyland and Hodin, 2004; Heyland and Moroz, 2005; Heyland et al., 2006a, b). Larvae of two species of abalone, Haliotis discus discus and H. gigantea metamorphosed strongly in response to T4. The metamorphic response to T4 was comparable to the response to gammer amino butyric acid (GABA), which is a highefficiency inducer of abalone metamorphosis (Fukazawa et al., 2001).
In mammals, THs (T4 and T3) are produced in the thyroid gland, and thyroid peroxidase (TPO) is the primary enzyme of THs biosynthesis (Degroot and Niepomniszcze, 1977; Kimura et al., 1987). In the target tissue, idothyronine deiodinases (ID) convert T4 to T3 (Flatt et al., 2006; Orozco et al., 2012). Cellular actions of THs may be initiated by two different ways. THs bind thyroid hormone receptors (TRs) and regulate gene expression depending on thyroid hormone response elements (TREs). This process happens in the cell nucleus and is termed genomic mechanism (Cheng et al., 2010). Moreover, via the integrin receptor, THs can regulate signal pathways, such as extracellular signal regulated kinases (ERK) (Lei et al., 2008; Lin et al., 2008; Cheng et al., 2010) or phosphatidylinositol 3-kinase (PI3K) (Lei et al., 2004; Cheng et al., 2010). Activation of ERK or PI3K involves TRs resident in cytoplasm (Cheng et al., 2010). This process is termed nogenomic mechanism. TRs play an important role in both ways, and therefore the biological functions of THs are mediated largely through TRs.
Vertebrate's TRs regulate growth, development and metabolism by binding either as monomers, homodimers or form heterodimers with RXRs (Muñoz et al., 1993). However, the function of TRs in invertebrates is still unknown. TRs have been reported in limited invertebrate groups. For example, a TR has been characterized in amphioxus (Paris et al., 2008). As well, an oyster TR ortholog has been identified (Huang et al., 2015). Moreover, there are TR orthologs in several species of trematode (Bertrand et al., 2004; Wu et al., 2006, 2007; Pakharukova et al., 2014). However, there are few reports about the role of TRs in marine invertebrate metamorphosis.
The spiral-cleaving embryos of abalone develop into trochophore larvae that undergo torsion and become free-swimming, lecithotrophic (nonfeeding) veligers (Williams et al., 2009). Then, 3 to 4 days later, H. diversicolor veligers become competent and are able to be induced by signals released by diatoms, mucus of adult, or biofilm to settle and metamorphose (Bryan and Qian, 1998; Li et al., 2006). TH has been shown to induce settlement and metamorphosis in two species of abalones (Fukazawa et al., 2001). H. diversicolor is commercially important in the southeast coast of China as a primary cultured species. In this study, the homolog of TRs from H. diversicolor was cloned and its mRNA expression profiles in metamorphosis and RNAi experiments were determined. In order to analyze abalone TRs function in metamorphosis, the expression profiles of TRrelated genes were showed in RNAi experiments and T3 inducing experiments. The understanding of TRs in metamorphosis will enhance our knowledge about life history of marine invertebrate, and contribute to seeding production of abalone.
2 MATERIAL AND METHOD 2.1 Larval sampleMature H. diversicolor and larval culture were carried out at Hongyun Aquaculture Co. Ltd. Abalone larvae were obtained as described in our previous paper (Wang et al., 2016). In brief, fertilized eggs and larvae were cultured in sand-filtered seawater at 23– 25℃. The larvae developed into veliger at 48 hours post fertilization (hpf), competent larva stage at 72 hpf, and post larva stage at 96 hpf. Six broods of larvae from different parents were collected into liquid nitrogen at veliger stage (48 hpf), competent larva stage (72 hpf) and post larva stage (96 hpf). Each brood larvae were derived from gametes contributions from at least three males and three females. Some competent larvae were used in dsRNA exposure assay and T3 inducing assay. When larva develops two eyespots, it is competent for metamorphosis, and becomes competent larva. When 80% larvae developed their eyespots, we consider these larvae as competent.
2.2 Isolation of total RNA and synthesis of cDNATotal RNA was extracted as described in our published peer-reviewed article (Wang et al., 2016). The total RNA was examined by agarose gel electrophoresis and ultraviolet spectrophotometer to ascertain its integrity and quantity. Then the total RNA was treated with RQ1 RNase-Free Dnase (Promega, Beijing, China) to remove potential trace amount of contaminating DNA. The cDNA was synthesized with 2 μg total RNA+0.5 μmol/L random primer+M-MLV reverse transcriptase, that was described in our published peer-reviewed article (Wang et al., 2016).
2.3 Molecular cloning of HdTRBLAST analysis of our transcriptome of H. diversicolor larvae revealed that a partial sequence was homologous to known sequences of TR. Gene special primers were designed based on the partial sequence (Table 1). RACE was performed to obtain the full length sequence of HdTR. SMART RACE Kit (Clontech, Mountain View, CA, USA) was used to get the 5′ and 3′ end according to its instructions. Expected PCR products were isolated by agarose gel electrophoresis and purified with a Qiaquick Gel Extraction Kit (Qiagen, Hilden, Germany). The purified DNA was ligated into the T/A cloning vector pMD19-T (TaKaRa, Dalian) to construct a recombinant vector, which was transfected into E. coli JM109 competent cells. The positive clones were sent to Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (China) to sequence using ABI 3730 automated sequencers (Applied Biosystems, Foster City, CA, USA).
Isoelectric point and molecular weight were computed using 'Compute pI/Mw tool' (http://cn.expasy.org/tools/pi_tool.html). Potential N-glycosylation and phosphorylation sites were analyzed with NetNGlyc1.0 Server (http://www.cbs.dtu.dk/). Protein multiple-alignments were performed with the BioEdit program (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The phylogenetic tree of TRs was constructed using maximum likelihood method with MEGA7 program (http://www.megasoftware.net/). Retinoic acid receptor of H. diversicolor was used as an outgroup.
2.4 dsRNA generationThe fragment of HdTR (521–1 056 bp of KT023066) was added T7 promoter sequence via PCR using dsTRFT7 and dsTR-R primers or using dsTR-F and dsTRRT7 primers. A 657-bp fragment of the green fluorescent protein (GFP) gene from the pEGFP-N1 vector was also added T7 promoter sequences by eithere dsGFP-FT7 and dsGFP-R or dsGFP-F and dsGFP-RT7. The sequences of primers were showed in Table 1. The italics at 5′ ends are T7 promoter sequence.
After purification and sequencing, the DNA fragments of HdTR and GFP were transcribed to single-stranded RNA (ssRNA) by T7 phage RNA polymerases (Promega, Fitchburg, USA). The DNA templates were degraded by DNase I (Promega, Fitchburg, USA). After purified, the sense and antisense cRNA strands were used to produce dsRNA, which was monitored by agarose gel electrophoresis and spectrophotometrically. The dsRNA of HdTR were used to silence HdTR. GFP dsRNA was served as the control. The details of dsRNA generation were described in our previous paper (Wang et al., 2016).
2.5 dsRNA exposure assayCompetent larvae were collected by 100 μm screens, and washed with 0.2 μm filtered seawater (FSW) into 50 mL beakers. The HdTR dsRNA/GFP dsRNA was added in a 50-mL beaker with a final concentration of 5 μg/mL. The beaker with HdTR dsRNA/GFP dsRNA was regarded as RNAi group / negative control (NC) group. The beaker without any dsRNA was regarded as blank control (BC) group. After 2 h, larvae were transferred into a 1-L beaker, which was covered by benthic diatom. Larvae in beaker were collected after 10 h transferred. Approximately 50 larvae were used to count the number of metamorphosed and dead larvae. Metamorphosis is defined as losing cilia and forming juvenile shell. Death is defined as deterioration of the outer cuticle and inactivity (Bryan and Qian, 1998). Larvae of different development stages were put into liquid nitrogen for RNA isolation then real time qRTPCR assay. The expression level of HdTR, TPO, DIII and effector genes (CCND1, CTNNB, ERK and PI3K) of THs were detected by qRT-PCR. The sequences of these genes were selected from transcriptomes (Huang et al., 2012) and ESTs (Jiang et al., 2011) of H. diversicolor. All sequences of transcritptomes and ESTs were aligned with Nr and Swissprot database by BLAST. The BLAST results showed that the sequences of our manuscript had the highest scores and lowest e-value with TPO, IDIII, CCND1, CTNNB, ERK and PI3K of other species, respectively. We had amplified these sequences by PCR and verified by Sanger's sequencing method. There were six replicate beakers of each treatment. After normality and homogeneity, data were analyzed with a one-way ANOVA and Fisher's LSD test to compare difference among treatments.
2.6 The assay of T3 inducing metamorphosisCompetent larvae were used in the inducing assay. A stock solution of T3 was prepared in FSW at a concentration of 100 μmol/L. Competent larvae were exposed in different T3 assay concentrations of 10, 1 and 0.1 μmol/L. The FSW without adding T3 was regarded as blank control. In detail, larvae collected by screening were put into 1-L beaker covered by benthic diatom with different T3 concentration. After 12 h, larvae were collected for metamorphosis rate counting and the sample collections of real time quantitative PCR (qRT-PCR) and whole mount in situ hybridization (WISH), which were similar to description in dsRNA exposure assay.
2.7 qRT-PCRThe detection of gene expression war carried out according to our previous report (Li et al., 2012; Wang et al., 2016). Briefly, total RNA was extracted from the samples with TRIZOL Reagent (Invitrogen, USA), and cDNA was generated by M-MLV (Promega, Fitchburg, USA). qRT-PCR was performed with an ABI 7500 real-time system by SYBR Green. Y-box binding protein gene (YB-1) was used as a reference, because of its constant expression in development (Huang et al., 2012). The comparative threshold cycle method was used to calculate the relative concentration. Six sample from different treatments were tested (n=6) as larval replicates, and each sample was assayed in triplicate for technical replicates according ABI qRT-PCR guide. The normalized CT values were analyzed by One-way ANOVA and Duncan variant significance tests. The genes and primers used in quantitative PCR were listed in Table 1.
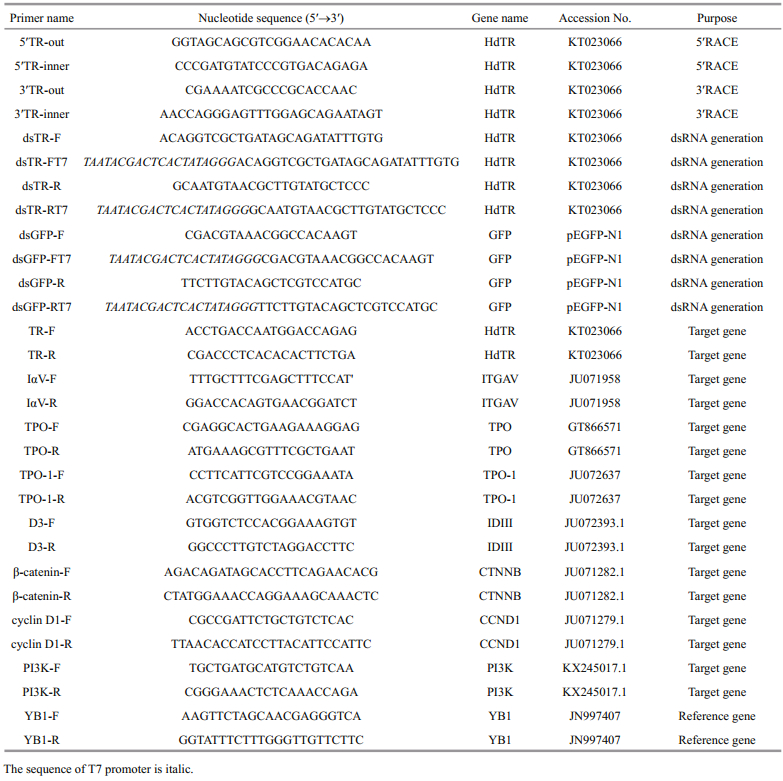
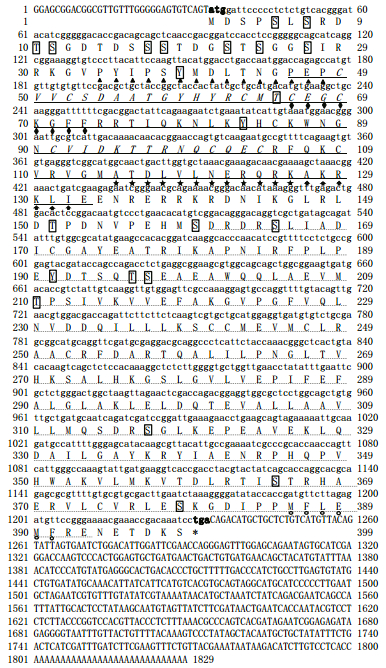
3 RESULT 3.1 Identification and characterization of a fulllength HdTR cDNA from abaloneThe full-length cDNA of HdTR is 1 829 bp, with 33 bp 5′ un-translated region (UTR) and 596 bp 3′ UTR (Fig. 1). The open reading frame is comprised of 1 200 bp, which encodes a protein (HdTR) of 399 amino acid residues. The calculated molecular mass of HdTR is 44.93 kDa, and the calculated isoelectric point is 8.40. Like typical nuclear receptor, HdTR contains a DBD and LBD (Fig. 1). Alignment of HdTR and other TR sequences showed that DBD of HdTR is the most conserved domain, which contains two zinc finger motifs (Figs. 1, 2). P-box sequence is highly conserved, which determines the specificity of target DNA binding. The T-box and A-box are also conserved. NTSS sequence of HdTR is PYIPSYMDLTNGPEP, and is conserved in nonchordate. While in chordates, three amino acids are deleted generating a sequence of GYIPSYL(D/E) KDE(P/Q/L) (Fig. 2). The analysis of phylogenetic relationship indicated that TRs were divided into two branches, one comprising vertebrate, and the other one containing invertebrate. H. diversicolor and three bivalve species formed a cluster, and a starfish and acorn worm formed another one. The two clusters and a cluster of three amphioxus species formed the invertebrate branch, which corresponded with the evolutionary relationships among these invertebrate species (Fig. 3).
|
| Fig.1 The characters of HdTR cDNA sequence and its encoding protein sequence The "*" is terminator, and the initiation codon, termination codon and tail signal is in bold. The box is on behalf of phosphorylation site. Underline and dashed part respectively indicate DNA binding domain (DBD domain) (46aa–133aa) and ligand binding domain (LBD domain) (161aa–393aa). Zinc finger structure is in italic. N-terminal signature sequence (NTSS, 34aa- 48aa) P-box (66aa– 73aa), T-box (115aa–126aa), A-box (127aa–132aa) and AF2 (385aa–391aa) are labeled by triangle, rhombus, star and arrow, respectively. |
|
| Fig.2 Multiple sequence alignment of HdTR amino acid sequence with other known TR proteins The identity and similar amino acid residues are highlighted in black and gray, respectively. LDB domain is labeled by blue line and DBD by purple line. The conserved NTSS, P-box, T-box, A-box and AF2 are framed in red. The accession numbers of TR protein sequences are: Haliotis diversicolor (ALM96676.1), Crassostrea gigas (AKE80988.1), Capitella teleta (ELT99607.1), Lingula anatine (XP_013406710.1), Saccoglossus kowalevskii (XP_006822150.1), Branchiostoma floridae (ABS11249.1), Necturus maculosus (AAO62999.1), Xenopus tropicalis (ADL74876.1), Homo sapiens (NP_000452.2), Danio rerio (NP_571415.1). |

|
| Fig.3 Phylogenetic tree of HdTR and other known TR Retinoic acid receptor of H. diversicolor, marked with triangle, is used as an outgroup. HdTR is marked with star. The GenBank accession No. of each TR is shown with its species name. |
The relative expression level of HdTR to YB1 is showed in Fig. 4. There was a significant difference of HdTR expression the three in the detected three stages (P=9.86E-11). HdTR expression of competent larva was significant lower than veliger (P=0.042 25), and higher than post larva (P=0.000 391). TPO expression level rapidly declined from veliger stage to competent larva stage (P=0.026 52), but there was no significant difference between competent larva and post-larva (P=0.252 9). TPO-1 had only significant difference between competent larva and post-larva (P=0.026 16). IDIII and ITGAV of post-larva were significantly higher than those of veliger and competent larva (P=0.000 92 and 0.041 33 respectively).
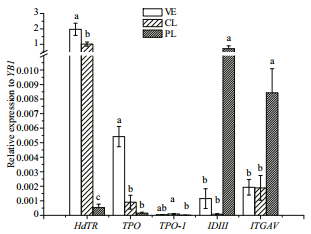
|
| Fig.4 The relative expressions of HdTR, TPO, TPO-1, IDIII and ITGAV were detected during metamorphosis YB1 served as reference gene. Each bar represents the mean value from six samples (n=6) with the standard error (SE). Three stages: veliger (VE), competent larvae (CL) and post larvae (PL) were detected by qPCR. The bars of each gene, if marked with different lowercase, indicate they are significantly different. The exact P values are described in the result section. |
After dsRNA exposure, the metamorphosis rate of RNAi group was significantly lower than that of BC and NC (P=0.042 64) (Fig. 5a). The mortality rate of RNAi group was significantly higher than that of BC and NC (P=0.001 0) (Fig. 5a). There were no significant differences of metamorphosis rate and mortality rate between BC and NC (P=0.069 99, P=0.072 29 respectively).

|
| Fig.5 Metamorphosis rate, mortality and genes expressions in dsRNA exposure Each bar represents the mean value from six samples (n=6) with the standard error (SE). The bars of metamorphosis rate or mortality in (a), if marked with different lowercase, indicate they are significantly different. The bars of each gene in (b), if marked with different lowercase, indicate they are significantly different. The exact P values are described in the result section. |
The exposure of HdTR dsRNA significantly reduced the expression level of HdTR (P=0.043 79) (Fig. 5b). HdTR expression dropped from 1.39 of NC group to 0.41 of RNAi group (P=0.014 67), and was inhibited 70.26%. The HdTR expression of BC was 0.97, and 57.38% of HdTR expression was inhibited with RNAi group (P=0.029 97). There were no significant expression differences of TPO, TPO-1 and IDIII, which encode the key enzymes of thyroid synthesis and conversion (Fig. 5b).
In order to determine the signaling pathway of HdTR, genes of genomic signaling pathway and nongenomic signaling pathway were detected by qRT-PCR. In nongenomic signaling pathway, ITGAV of RNAi group was significantly lower than that of BC and NC (P=0.042 23, Fig. 5b). The expression levels of other genes had no significant changes in the exposure of dsRNA HdTR (P=0.874 6, P=0.856 0, Fig. 5b). In genomic signaling pathway, CTNNB of dsRNA HdTR was higher than that of BC and NC (P=0.039 62). CCND1 of RNAi group were lower than that of BC and NC (P=0.034 76, Fig. 5b).
3.4 Effect of T3 inducing on metamorphosis rate and genes expressionT3 had a significant effect on metamorphosis rate and mortality (P=0.037 697 and 0.028 466 respectively) (Fig. 6a). The metamorphosis rate of 1 μmol/L T3 was higher than the other concentrations of T3 (P=0.037 697), while the mortality of 1 μmol/L T3 was lower than the other concentration of T3 (P=0.028 466). But the metamorphosis rate and mortality rate of 10 μmol/L T3 and 0.1 μmol/L T3 had no significant difference compared to that of 0 μmol/L T3 (P=0.333 704, 0.606 512; and 0.672 621, 0.290 906 respectively).

|
| Fig.6 Metamorphosis rate, mortality and genes expressions in T3 exposure Each bar represents the mean value from six samples (n=6) with the standard error (SE). The bars of metamorphosis rate or mortality in (a), if marked with different lowercase, indicate they are significantly different. The bars of each gene in (b), if marked with different lowercase, indicate they are significantly different. The exact P values are described in the result section. |
There was no significant difference of TPO/TPO-1 expression in different T3 concentrations (P=0.170 1 and 0.910 5, respectively) (Fig. 6b). IDIII expression of 0 μmol/L T3 had no significant difference than other T3 concentrations (P=0.328 3, 0.115 5 and 0.524 2, respectively). In nongenomic signaling pathway, ITGAV expression of 1 μmol/L T3 and 0.1 μmol/L T3 was lower than that of 0 μmol/L T3 (P=0.036 678 and 0.033 265 respectively) (Fig. 6b). But PI3K expression of 10 μmol/L T3 was higher than that of 0 μmol/L T3 (P=0.046 908). ERK expressions of different T3 concentrations had no significant difference (P=0.236 5). In genomic signaling pathway, HdTR expressions of 1 μmol/L T3 and CCND1 expressions of 0.1 μmol/L T3 were higher than that of 0 μmol/L T3 (P=0.032 877 and 0.046 99, respectively) (Fig. 6b). CTNNB expressions had no significant difference in different T3 concentrations (P=0.304 3).
4 DISCUSSIONNuclear receptors are exclusive to multicellular metazoans. As a member of nuclear receptor (NR) gene family, TR was previously believed to be an innovation of chordates as no TR orthologs have been identified in the genomes of insects and nematodes (Bertrand et al., 2004). The presence of TR homologues in several invertebrates demonstrated that the TR orthologue genes are present outside of chordates. They were considered to originate in a common ancestor of bilateria (Wu et al., 2007). Although the N-terminal of NR is highly divergent, there is a conserved NTSS (N-terminal signature sequence), which is TR-specific. TR has two highly conserved domains, which are DBD and LBD (Wu et al., 2007). The TRs of flatworm Schistosoma mansoniare highly conserved not only in sequence similarity, but also in gene organization, proteinprotein interaction and in DNA-binding ability. This suggests that the sequence and function of TRs are highly conserved between invertebrate animals (but not Porifera or Cnidaria) and vertebrate animals (Wu et al., 2007). HdTR has conserved sequence, including NTSS, DBD and LBD. This suggests that HdTR may have similar function of vertebrate TR.
THs are small, lipophilic signaling molecules built from tyrosine and iodine. THs primary source of marine invertebrate is marine phytoplankton, although some larvae may synthesize THs (Heyland and Moroz, 2005). THs may be transferred through the food chain and work as a cross-kingdom hormonal signaling (Heyland and Moroz, 2005). Heyland and Hodin (2004) had shown that THs can act via exogenous routes as environmental messengers in echinoderm larvae. Abalone larvae (H. discus discus and H. gigantea) were induced metamorphosis by THs at the concentration of 1 μmol/L (Fukazawa et al., 2001). Our findings are similar to this paper. The T3 inducing abalone metamorphosis is similar to the case in echinoderm larvae. T3 may take a message of metamorphosis, which means environment is suitable for metamorphosis of abalone larvae.
There was a high expression level of HdTR in swimming larvae stage and a rapid decline at post larva stage. When the high expression of HdTR was reduced by RNAi at competent larvae stage, there appeared lower metamorphosis rate and higher mortality rate. Moreover, the expression pattern of HdTR was similar to that of TPO and TPO-1. This means that HdTR expression level has positive correlation with endogenous THs. And the significant increase of HdTR expression is associated with increase of metamorphosis rate in 1 μmol/L T3. These data suggest that HdTR is involved in inducing metamorphosis of THs.
It is well known that TR binding THs can regulate gene expression with TREs. It is termed genomic action of THs. CCND1 and CTNNB are effective genes of genomic signaling pathway (Cheng et al., 2010). The expression of these four genes had a significant difference in HdTR RNAi, but their expression patterns of 0.1 μmol/L T3 were different. Only CCND1 had a higher expression in 0.1 μmol/L T3 than that in control (0 μmol/L T3). The expressions of CTNNB in 0.1 μmol/L T3 had no significant difference compared to that in 0 μmol/L T3. This result showed that TR genomic action may only mediate the expression of CCND1. CCND1 encodes the cyclin D1 protein, which is required for progression through the G1 phase of the cell cycle (Baldin et al., 1993). CCND1 is an early target in hepatocyte proliferation of rat induced by the THs (Pibiri et al., 2001), and TR deficient mice exhibit a significant delay in liver cell proliferation (Pascual and Aranda, 2013). These results are similar to our findings. Abalone larvae need develop juvenile/adult structures (such as gill, foot, and digestive system) in metamorphosis (Bishop, 2006; Heyland and Moroz, 2006). The cell proliferation is exuberant in the organogenesis. The up-regulated expression of CCND1 suggests that the effect of TR genomic action may mainly show in cell proliferation during metamorphosis.
The transcriptional activity of the TRs is modulated by T3, but there is also activity in the unliganded state (Bernal and Morte, 2013). The unliganded TRs interact with the corepressors NCoR (nuclear receptor corepressor) and SMRT (silencing mediator of retinoid and thyroid hormone receptor) and may repress or enhance gene expression (Cheng et al., 2010). The unliganded TRs are responsible for changes of many THs responsive genes in mouse liver (Yen et al., 2003). Meanwhile, the unliganded TRs influence the timing and control certain aspects of amphibian pre-metamorphosis (Sato et al., 2007). The expression changes of CTNNB in HdTR RNAi may be due to the activity of the unliganded TRs. CTNNB encodes β-catenin, a structural component of cell adhesion complexes, which regulates actin filament assembly to regulate cellular functions (Gottardi and Gumbiner, 2001). In addition, β-catenin also functions as a co-activator of transcription factors, and is involved in cell proliferation, survival, and migration (Moon et al., 2002). Our data suggested that CTNNB may play a role in new structure development of metamorphosis by unliganded HdTR.
T3 and other THs can also act via nongenomic signaling. These effects are mediated via membrane receptors, such as integrin αvβ3 (Bergh et al., 2005). ITGAV expression in 0.1 μmol/L T3 was lower than that in 0 μmol/L T3. This suggests that T3 may have a negative regulation on the expression of ITGAV. Via the integrin αvβ3, T3 stimulates ERK through phospholipase C (PLC) and protein kinase C (PKC). The signaling process is based on protein phosphorylation. The mRNA expression of ERK was not significantly different. The results did not suggest that integrin αvβ3 signal pathway does not work. However, we had not detected the protein activity of ERK and PI3K. As the cascade effect of αvβ3 is dependent on the protein activity of molecular components in the signal pathway, we are not sure that integrin αvβ3 signal pathway works, neither.
The Na, K-ATPcase activity stimulated by T3 nongenomic effect requires PI3K. This means that PI3K is involved in another nongenomic signal pathways of T3 (Lei et al., 2004). The expression level of PI3K had a significant difference in T3 inducing, but no in HdTR RNAi. This means PI3K were affected by T3. Our data is consistent with these previous papers.
Metamorphosis is a life-history transition that involves radical changes in habitat, morphology, and physiology (Bishop et al., 2006). For abalone, the swimming larvae change to creeping juveniles. Then the larval velum is lost. Meanwhile, the juvenile gill, foot and secondary shell appear. The cell differentiation and apoptosis are involved in these processes. Moreover, metamorphosis is tightly regulated by hormones and a variety of environmental signals (Bishop et al., 2006). Many kinds of chemical and biological materials are used to induce metamorphosis of abalone larvae (Roberts et al., 2001). This suggests that there are massive signal pathways involving abalone metamorphosis and there are complicated interactions among these pathways. However, there are short of protein tools and materials in pathway analysis of abalone metamorphosis. We only try to analyze the role of TR depending on gene expression data. Our data showed that HdTR is involved in cell proliferation of metamorphosis by TR genomic action or unliganded TR effect, and energy metabolism by unliganded TR effect. T3 may induce some THs nongenomic effect, but we have no evidence in protein level.
5 CONCLUSIONIn summary, there is a significant decrease of metamorphosis rate after competent larvae were exposed to dsRNA of HdTR, and a significant increase in 1 μmol/L T3 inducing. The results suggested that HdTR and T3 play roles in abalone metamorphosis. The different expression patterns of thyroid hormone effector genes (CCND1, CTNNB, GSK, STAT1 and PI3K) in RNAi of HdTR and T3 inducing suggested that HdTR and T3 regulate metamorphosis by different ways. HdTR may be involved in cell proliferation of metamorphosis by TR genomic action or unliganded TR effect, and energy metabolism by unliganded TR effect in metamorphosis. T3 may induce some THs nongenomic effect in metamorphosis.
6 DATA AVAILABILITY STATEMENTHdTR had been deposited in the GenBank database under accession number KT023066.1.
Baldin V, Lukas J, Marcote M J, Pagano M, Draetta G. 1993. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes & Development, 7(5): 812-821.
|
Bergh J J, Lin H Y, Lansing L, Mohamed S N, Davis F B, Mousa S, Davis P J. 2005. Integrin αVβ3 contains a cell surface receptor site for thyroid hormone that is linked to activation of mitogen-activated protein kinase and induction of angiogenesis. Endocrinology, 146(7): 2864-2871.
DOI:10.1210/en.2005-0102 |
Bernal J, Morte B. 2013. Thyroid hormone receptor activity in the absence of ligand:physiological and developmental implications. Biochimica et Biophysica Acta (BBA)-General Subjects, 1830(7): 3893-3899.
DOI:10.1016/j.bbagen.2012.04.014 |
Bertrand S, Brunet F G, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. 2004. Evolutionary genomics of nuclear receptors:from twenty-five ancestral genes to derived endocrine systems. Molecular Biology and Evolution, 21(10): 1923-1937.
DOI:10.1093/molbev/msh200 |
Bishop C D, Erezyilmaz D F, Flatt T, Georgiou C D, Hadfield M G, Heyland A, Hodin J, Jacobs M W, Maslakova S A, Pires A, Reitzel A M, Santagata S, Tanaka K, Youson J H. 2006. What is metamorphosis?. Integrative and Comparative Biology, 46(6): 655-661.
DOI:10.1093/icb/icl004 |
Bryan P J, Qian P Y. 1998. Induction of larval attachment and metamorphosis in the abalone Haliotis diversicolor(Reeve). Journal of Experimental Marine Biology and Ecology, 223(1): 39-51.
DOI:10.1016/S0022-0981(97)00156-1 |
Cheng S Y, Leonard J L, Davis P J. 2010. Molecular aspects of thyroid hormone actions. Endocrine Reviews, 31(2): 139-170.
|
Degroot L J, Niepomniszcze H. 1977. Biosynthesis of thyroid hormone:basic and clinical aspects. Metabolism, 26(6): 665-718.
DOI:10.1016/0026-0495(77)90088-9 |
Flatt T, Moroz L L, Tatar M, Heyland A. 2006. Comparing thyroid and insect hormone signaling. Integrative and Comparative Biology, 46(6): 777-794.
DOI:10.1093/icb/icl034 |
Fukazawa H, Hirai H, Hori H, Roberts R D, Nukaya H, Ishida H, Tsuji K. 2001. Induction of abalone larval metamorphosis by thyroid hormones. Fisheries Science, 67(5): 985-988.
DOI:10.1046/j.1444-2906.2001.00351.x |
Gottardi C J, Gumbiner B M. 2001. Adhesion signaling:how β-catenin interacts with its partners. Current Biology, 11(19): R792-R794.
DOI:10.1016/S0960-9822(01)00473-0 |
Heyland A, Hodin J. 2004. Heterochronic developmental shift caused by thyroid hormone in larval sand dollars and its implications for phenotypic plasticity and the evolution of nonfeeding development. Evolution, 58(3): 524-538.
DOI:10.1111/evo.2004.58.issue-3 |
Heyland A, Moroz L L. 2005. Cross-kingdom hormonal signaling:an insight from thyroid hormone functions in marine larvae. Journal of Experimental Biology, 208(Pt 23): 4355-4361.
|
Heyland A, Moroz L L. 2006. Signaling mechanisms underlying metamorphic transitions in animals. Integrative and Comparative Biology, 46(6): 743-759.
DOI:10.1093/icb/icl023 |
Heyland A, Price D A, Bodnarova-Buganova M, Moroz L L. 2006a. Thyroid hormone metabolism and peroxidase function in two non-chordate animals. Journal of Experimental Zoology, 306B(6): 551-566.
DOI:10.1002/jez.b.v306b:6 |
Heyland A, Reitzel A M, Price D A, Moroz L L. 2006b. Endogenous thyroid hormone synthesis in facultative planktotrophic larvae of the sand dollar Clypeaster rosaceus:implications for the evolutionary loss of larval feeding. Evolution & Development, 8(6): 568-579.
|
Holstein T W, Laudet V. 2014. Life-history evolution:at the origins of metamorphosis. Current Biology, 24(4): R159-R161.
DOI:10.1016/j.cub.2014.01.003 |
Huang W, Xu F, Qu T, Zhang R, Li L, Que H Y, Zhang G F. 2015. Identification of thyroid hormones and functional characterization of thyroid hormone receptor in the pacific oyster Crassostrea gigas provide insight into evolution of the thyroid hormone system. PLoS One, 10(12): e0144991.
DOI:10.1371/journal.pone.0144991 |
Huang Z X, Chen Z S, Ke C H, Zhao J, You W W, Zhang J, Dong W T, Chen J. 2012. Pyrosequencing of Haliotis diversicolor transcriptomes:insights into early developmental molluscan gene expression. PLoS One, 7(12): e51279.
DOI:10.1371/journal.pone.0051279 |
Jiang J Z, Zhang W, Guo Z X, Cai C C, Su Y L, Wang R X, Wang J Y. 2011. Functional annotation of an expressed sequence tag library from Haliotis diversicolor and analysis of its plant-like sequences. Marine Genomics, 4(3): 189-196.
|
Johnson L G. 1998. Stage-dependent thyroxine effects on sea urchin development. New Zealand Journal of Marine and Freshwater Research, 32(4): 531-536.
DOI:10.1080/00288330.1998.9516841 |
Kimura S, Kotani T, McBride O W, Umeki K, Hirai K, Nakayama T, Ohtaki S. 1987. Human thyroid peroxidase:complete cDNA and protein sequence, chromosome mapping, and identification of two alternately spliced mRNAs. Proceedings of the National Academy of Sciences of the United States of America, 84(16): 5555-5559.
DOI:10.1073/pnas.84.16.5555 |
Lei J X, Mariash C N, Bhargava M, Wattenberg E V, Ingbar D H. 2008. T3 increases Na-K-ATPase activity via a MAPK/ERK1/2-dependent pathway in rat adult alveolar epithelial cells. American Journal of Physiology-Lung Cellular and Molecular Physiology, 294(4): L749-L754.
DOI:10.1152/ajplung.00335.2007 |
Lei J X, Mariash C N, Ingbar D H. 2004. 3, 3', 5-Triiodo-lthyronine up-regulation of Na, K-ATPase activity and cell surface expression in alveolar epithelial cells is Src kinase-and phosphoinositide 3-kinase-dependent. Journal of Biological Chemistry, 279(46): 47589-47600.
DOI:10.1074/jbc.M405497200 |
Li H F, Lin W, Zhang G, Cai Z H, Cai G P, Chang Y Q, Xing K Z. 2006. Enhancement of larval settlement and metamorphosis through biological and chemical cues in the abalone Haliotis diversicolor supertexta. Aquaculture, 258(1-4): 416-423.
DOI:10.1016/j.aquaculture.2006.04.013 |
Li N, Zhang Z P, Zhang L L, Wang S H, Zou Z H, Wang G D, Wang Y L. 2012. Insulin-like growth factor binding protein 7, a member of insulin-like growth factor signal pathway, involved in immune response of small abalone Haliotis diversicolor. Fish & Shellfish Immunology, 33(2): 229-242.
|
Lin H Y, Tang H Y, Keating T, Wu Y H, Shih A, Hammond D, Sun M Z, Hercbergs A, Davis F B, Davis P J. 2008. Resveratrol is pro-apoptotic and thyroid hormone is antiapoptotic in glioma cells:both actions are integrin and ERK mediated. Carcinogenesis, 29(1): 62-69.
|
Moon R T, Bowerman B, Boutros M, Perrimon N. 2002. The promise and perils of Wnt signaling through β-catenin. Science, 296(5573): 1644-1646.
DOI:10.1126/science.1071549 |
Muñoz A, Wrighton C, Seliger B, Bernal J, Beug H. 1993. Thyroid hormone receptor/c-erbA:control of commitment and differentiation in the neuronal/chromaffin progenitor line PC12. Journal of Cell Biology, 121(2): 423-438.
|
Orozco A, Valverde-R C, Olvera A, Garcia-G C. 2012. Iodothyronine deiodinases:a functional and evolutionary perspective. Journal of Endocrinology, 215(2): 207-219.
|
Pakharukova M Y, Ershov N I, Vorontsova E V, Shilov A G, Merkulova T I, Mordvinov V A. 2014. Identification of thyroid hormone receptor homologs in the fluke Opisthorchis felineus (platyhelminthes). Molecular and Biochemical Parasitology, 194(1-2): 64-68.
DOI:10.1016/j.molbiopara.2014.04.009 |
Paris M, Escriva H, Schubert M, Brunet F, Brtko J, Ciesielski F, Roecklin D, Vivat-Hannah V, Jamin E L, Cravedi J P, Scanlan T S, Renaud J P, Holland N D, Laudet V. 2008. Amphioxus postembryonic development reveals the homology of chordate metamorphosis. Current Biology, 18(11): 825-830.
DOI:10.1016/j.cub.2008.04.078 |
Pascual A, Aranda A. 2013. Thyroid hormone receptors, cell growth and differentiation. Biochimica et Biophysica Acta(BBA)-General Subjects, 1830(7): 3903-3916.
|
Pibiri M, Ledda-Columbano G M, Cossu C, Simbula G, Menegazzi M, Shinozuka H, Columbano A. 2001. Cyclin D1 is an early target in hepatocyte proliferation induced by thyroid hormone (T3). The FASEB Journal, 15(6): 1006-1013.
DOI:10.1096/fj.00-0416com |
Roberts R D, Lapworth C, Barker R J. 2001. Effect of starvation on the growth and survival of post-larval abalone (Haliotis iris). Aquaculture, 200(3-4): 323-338.
DOI:10.1016/S0044-8486(01)00531-2 |
Sato Y, Buchholz D R, Paul B D, Shi Y B. 2007. A role of unliganded thyroid hormone receptor in postembryonic development in Xenopus laevis. Mechanisms of Development, 124(6): 476-488.
DOI:10.1016/j.mod.2007.03.006 |
Wang G D, Li N, Zhang L L, Zhang L H, Zhang Z P, Wang Y L. 2016. IGFBP7 is involved in abalone metamorphosis. Aquaculture, 451: 377-384.
DOI:10.1016/j.aquaculture.2015.09.031 |
Williams E A, Degnan B M, Gunter H, Jackson D J, Woodcroft B J, Degnan S M. 2009. Widespread transcriptional changes pre-empt the critical pelagic-benthic transition in the vetigastropod Haliotis asinina. Molecular Ecology, 18(5): 1006-1025.
DOI:10.1111/mec.2009.18.issue-5 |
Wu W J, Niles E G, El-Sayed N, Berriman M, Loverde P T. 2006. Schistosoma mansoni (Platyhelminthes, Trematoda)nuclear receptors:sixteen new members and a novel subfamily. Gene, 366(2): 303-315.
DOI:10.1016/j.gene.2005.09.013 |
Wu W J, Niles E G, Hirai H, Loverde P T. 2007. Evolution of a novel subfamily of nuclear receptors with members that each contain two DNA binding domains. BMC Evolutionary Biology, 7: 27.
DOI:10.1186/1471-2148-7-27 |
Yen P M, Feng X, Flamant F, Chen Y D, Walker R L, Weiss R E, Chassande O, Samarut J, Refetoff S, Meltzer P S. 2003. Effects of ligand and thyroid hormone receptor isoforms on hepatic gene expression profiles of thyroid hormone receptor knockout mice. EMBO Reports, 4(6): 581-587.
DOI:10.1038/sj.embor.embor862 |
 2019, Vol. 37
2019, Vol. 37