Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Huanshan, WANG Teng, LI Wenjing, LIU Huanzhang
- The genetic diversity, individual relatedness and possible mating system of an isolated population of the Cyprinid species Megalobrama pellgrini in upper reaches of the Changjiang (Yangtze) River, China
- Journal of Oceanology and Limnology, 37(3): 1042-1050
- http://dx.doi.org/10.1007/s00343-019-8152-7
Article History
- Received May. 11, 2018
- accepted in principle Jun. 19, 2018
- accepted for publication Jul. 11, 2018
2 University of Chinese Academy of Sciences, Beijing 100049, China
One of the greatest contributions for the new Darwinism is the recognition of the idea that all the species exists by the form of the population. To maintain high viability, a population has to be large in number and exchange genetic diversity with other populations (Huxley, 1940; Mayr, 1963). Once a population was isolated, it might experience a bottleneck effect (Allendorf and Luikart, 2007), suffer from inbreeding depression (Madsen et al., 1996; Allendorf and Luikart, 2007). This can lead to increasing linkage disequilibrium and decreasing allelic diversity in the population (Hauser et al., 2002; Pardo et al., 2005; Bonnen et al., 2006; Service et al., 2006; Bendjilali et al., 2014), result in decreased fitness and viability, and then ultimately lead the population to the so-called extinction vortex (Gilpin, 1986; Westemeier et al., 1998; Lindenmayer and Peakall, 2000; Grayson et al., 2014). Therefore, great concerns have been given to the isolated populations, some indicators have been proposed to evaluate the status of the populations, and many measures have been discussed for their conservation.
Megalobrama pellegrini is an endemic cyprinid fish in the upper reaches of the Changjiang (Yangtze) River (Luo, 1990; Chen, 1998; Li et al., 2007a). It has a very deep body depth in a bream shape and is very important economic fish in the local area. This fish used to inhabit widely the main channel and tributaries of the upper Changjiang River (Luo, 1990; Chen, 1998). However, in the recent decades, due to the intensive anthropologic activities like damming, pollution, and overfishing, it has become very rare in the main channel. Contrary to the resource scarcity in the main channel, M. pellegrini is found abundant in a tributary of the upper Changjiang River, the Longxi River (Li et al., 2007b). This river is approximately 97 km long with a drainage area of approximately 521 km2 and flows into the Changjiang River in Luzhou City, Sichuan Province. In lower reaches of this river, there is a natural cliff up to 35 m high, which forms a natural barrier isolating fish populations of this river from those of the main channel of the Changjiang River (Li et al., 2007b). The previous investigation showed that M. pellegrini population in the Longxi River has lower genetic diversity, and differentiated from the nearby population at the median level (Wang et al., 2014). Our recent investigations indicate that the M. pellegrini individuals in the Longxi River showed slower growth rate than it was 10 years ago (Li et al., 2007a), and the resources have also become diminished. Therefore, concerns have been raised on how the genetic status of this population is at, whether it has suffered from some genetic catastrophe, and how we should decide conservation measures for this population.
In the present study, we collected samples of M. pellegrini individuals from the Longxi River. Using two different molecular markers, mitochondrial cytochrome b gene and nuclear microsatellite (simple sequence repeat, SSR), we analyzed their genetic diversity with the purpose (1) to evaluate genetic status of this population and to reveal the probable causes, (2) to identify their mating system and individual relatedness, and (3) to give suitable conservation suggestions.
2 MATERIAL AND METHOD 2.1 Sampling120 individuals of M. Pellegrini were collected from the Longxi River in Luzhou, Sichuan Province by bottom gillnets (mesh size 4.0 cm) from July 2011 to June 2013. The tissues of muscle and fin were preserved in 95% ethanol.
2.2 Laboratory experimentsIn the laboratory, genomic DNA was extracted using salt-extraction method proposed by Aljanabi and Martinez (1997). Two different types of markers, cytochrome b (cyt b) gene of mitochondrial DNA (mtDNA), nuclear microsatellite (simple sequence repeat, SSR) were chosen and amplified by polymerase chain reaction (PCR), which were performed in volumes of 20 μL (SSR) or 30 μL (cyt b markers) containing 30–50 ng template DNA, 0.5 μL dNTP mixture (2.5 mmol/L each), 0.3 U Taq DNA Polymerase with MgSO4 (2 mmol/L of Mg2+), 2 μL 10×Taq Buffer, 0.5 μL each primer (10 μmol/L) and supplement by sterile ddH2O. The PCR reaction conditions for cyt b were: 94℃ for 4 min, then 35 cycles with 45 s at 94℃, 45 s at the annealing temperature and 1 min at 72℃, with a final elongation at 72℃ for 10 min; and for SSR were: 94℃ for 5 min, then 35 cycles with 30 s at 94℃, 40 s at the annealing temperature and 1 min at 72℃, with a final elongation at 72℃ for 10 min. The information of the primers is shown in Table 1. After being amplified, the PCR products were detected through agarose gel electrophoresis (cyt b) or 8% nondenaturing polyacrylamide gels (SSR). Then the fragments (cyt b) were sequenced with the same primers as for PCR by Shanghai Sangon Company while the gels (SSR) were stained with Ethidium bromide and viewed under UV light with Genesys software (Syngene).
cyt b: The alignment of cyt b sequences were performed with ClustalX version 2.0 (Larkin et al., 2007) and revised manually with SEAVIEW (Galtier et al., 1996). The diversity of population was analyzed using DnaSP v5.10 (Librado and Rozas, 2009) including the number of haplotypes (h), nucleotide diversity (Pi), haplotype diversity (Hd) and the average number of nucleotide differences (k). Software MEGA6.0 (Tamura et al., 2013) was used to calculate the nucleotide composition, conserved sites (C), variable sites (V), parsimony-informative sites (P), and singleton sites (S).
SSR: Genotypes were checked for scoring errors attributable to stutter-products, large allele dropout, or null alleles using Micro-Checker v2.2.3 (van Oosterhout et al., 2004). The number of alleles, allele size range, Ho (observed heterozygosity) and He (expected heterozygosity) were calculated using PopGene v1.32 (Yeh et al., 1997). Inbreeding coefficient (FIS) was estimated using Arlequin 3.5.1.2 (Excoffier and Lischer, 2010).
We estimated the effective population size (Ne) using the Linkage Disequilibrium Method, as implemented in NE-Estimator V2 (Do et al., 2014). The program BOTTLENECK v1.2.02 (Cornuet and Luikart, 1996) was used to test for evidence of recent bottleneck events based on theoretical expectations under the Stepwise Mutation Model (SMM) (Shriver et al., 1993; Valdes et al., 1993).
The pairwise distances between individuals were calculated by software Populations v1.2.32 (Langella, 2002) and the Neighbor-joining tree was built by MEGA6.0 (Tamura et al., 2013) based on the distance matrix. The relatedness between individuals was estimated by software ML-relate (Kalinowski et al., 2006) and COLONY version2.0.5.8 (Jones and Wang, 2010).
3 RESULT 3.1 Sequence genetic diversityWe got sequences of the complete cyt b gene (1 140 bp) for all the 120 samples. There were 9 haplotypes for cyt b gene. After alignment, there were no insertion and deletion.
The average nucleotide composition of cyt b gene were T=27.5%, C=29.0%, A=28.4%, and G=15.1% respectively (Table 2). The content of G was very low (15.1%), with the content of A+T (55.9%) much higher than that of G+C (44.1%). The Hd was 0.290 and Pi was 0.000 77 (Table 2). There were 11 variable sites (0.96%) including 6 parsimony-informative sites and 5 singleton sites (Table 2).
Null alleles were checked by Micro-Checker. There was no evidence of large allele dropout and scoring errors caused by stuttering, but through the 3 loci there existed null alleles (P < 0.05).
The expected and observed heterozygosity per locus ranged from 0.282 4 to 0.941 7 (He) and 0.178 0 to 1.000 0 (Ho), respectively (Table 3). The average levels of expected and observed heterozygosity were 0.823 5±0.145 1 (He) and 0.824 4±0.147 2 (Ho), respectively (Table 3). There were 12 loci with negative FIS and 8 loci with positive FIS. Polymorphism information content (PIC) per locus were from 0.259 6 to 0.938 7 with a mean of 0.805 7±0.155 0 (Table 3). HWE-test showed that 11 loci deviated from Hardy-Weinberg equilibrium.
The Wilcoxon test revealed the null hypothesis of mutation-drift equilibrium under SMM was significantly rejected (P=0.000 05 < 0.05), and this indicated a recent bottleneck.
Effective population size (Ne) of the whole population was calculated as 35.9, 52.7, and 70.5 respectively at different lowest allele frequency (lowest allele frequency used were 0.05, 0.02, and 0.01 correspondingly).
3.3 Individual relatednessIndividual relatedness for all the 120 samples was analyzed with the software ML-relate (Kalinowski et al., 2006) and COLONY version 2.0.5.8 (Jones and Wang, 2010). Their results were the same and showed that all the individuals were grouped into 10 completely independent clusters (Fig. 1). Among them, Cluster 3 and Cluster 4 had the most individuals and were relatively complex than the other ones (Fig. 1), while clusters 8, 9 and 10 only had 2 individuals respectively. This relatedness indicated that both male and female M. pellegrini might mate with several individuals showing a polygamy system.
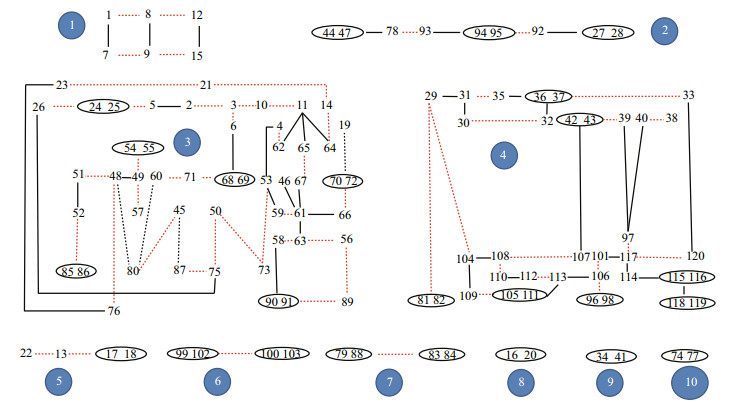
|
| Fig.1 The relatedness between 120 individuals Circles represent full-siblings, black line and red line represent the half-siblings with same father or same mother respectively; the numbers in blue circles represents the clusters they belong to. |
We also calculated the pairwise distances between individuals and then built a neighbor-joining tree based on the distance matrix (Fig. 2). The tree showed similar relationships to their relatedness clustered independently in overall but with several mixed samples.
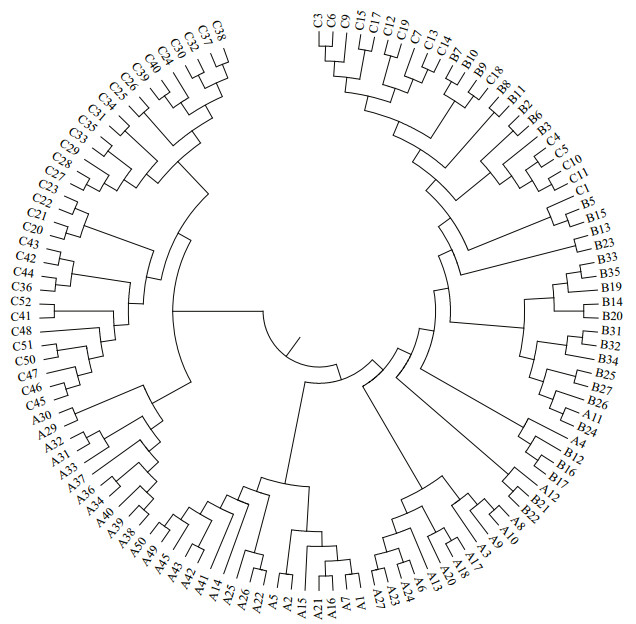
|
| Fig.2 The neighbor-joining tree based on the distance of DAS and Cp using 20 loci of microsatellite |
Normally animal-mating systems can be classified as two main types: mono-mating and poly-mating (Klug, 2011). It was suggested that the reproductive success of males is often limited by access to female mates, whereas the reproductive success of females is often limited by resource acquisition (Bateman, 1948). For the successful reproduction, males need females, and females need resources (Klug, 2011). Thus, the spatial distribution of resources may affect mating systems. It was hypothesized that single individuals may be difficult to monopolize more than one mate if critical resources are uniformly distributed in space. As a result, territoriality will occur, the potential for multiple mating will be low, and monogamy is likely to prevail (Emlen and Oring, 1977).
Alternatively, when critical resources are clumped, some proportion of individuals can potentially monopolize those resources, the potential for polygamy can be relatively high (Emlen and Oring, 1977). However, when the resources are highly clumped, it might become more difficult for individuals to monopolize resources due to the increased level of competition, and the potential for polygamy will possibly lessen (Klug, 2011).
Previous investigations show that M. pellegrini spawns adhesive eggs (Li et al., 2007b). In the present study, analysis to SSR data showed that M. pellegrini was in polygamy mode, indicating the spawning grounds of M. pellegrini were in clumped status.
4.2 Genetic diversity, bottleneck, and inbreedingSimilar to previous investigations, our present study showed that M. pellegrini population in the Longxi River was in low-level genetic diversity with cyt b. For mtDNA cyt b gene, the haplotype diversity was 0.290 and nucleotide diversity was 0.000 77, both were very low. Grant and Bowen (1998) suggested that this situation indicated a probable recent bottleneck for the population. Conversely, for SSR, the diversity was relatively high, with the average Ho and He as 0.824 4±0.147 2 and 0.823 5±0.145 1 respectively. Again, this low-level sequence diversity and high-level SSR diversity suggest the extremely recent population bottleneck (Lee et al., 2006). Population bottlenecks can increase rates of inbreeding, loss of genetic variation and fixation of mildly deleterious alleles, and thereby reduce adaptive potential and increase the probability of extinction (Cornuet and Luikart, 1996; Luikart et al., 1998).
Besides, inbreeding was also found in this population. There were 12 loci with negative FIS, indicating heterozygote excess, and 8 loci with positive FIS meaning the population existing inbreeding. Inbreeding may depress the reproductive fitness and growth rate, reduce genetic variability, viability, and fecundity, and result in accumulations of new deleterious mutations. In addition, inbreeding is considered a fatal factor that will increase the risk of extinction of a wild population (Keller et al., 1994; Frankham and Ralls, 1998; Daniels et al., 2000; Brook et al., 2002).
In the present study, the effective population size was estimated anywhere from 35.9 to 70.5 under different allele frequency, close to 50. When Ne is low, populations can be hard to maintain a long-term survival. It was suggested that the effective population size should be no less than 50 for short time survival and 500 for long-term survival (Franklin, 1980). The Ne of our investigated M. pellegrini population was calculated at around 50, which is at the lower limit. We suggested that the population size should be increased.
4.3 Conservation suggestionsAnalysis of the present study showed that M. pellegrini population in the Longxi River indeed have suffered from some genetic catastrophe, such as the extremely low genetic diversity, inbreeding, recent bottleneck and small effective population size. These results also indicated that the most urgent genetic catastrophe was not induced by the isolation, but overfishing and population miniaturization (Gao et al., 2009, Wang et al., 2014). As for the M. pellegrini, it lived in an isolating water environment and suffered from intensive artificial pressure, which may result in a small population size and increase the risk of extinction. Therefore, we suggested the urgent conservation measures should be taken to control fishing, so the fish can grow and reproduce in a normal mode, and then to restore the population size.
5 CONCLUSIONIn conclusion, we selected two molecular markers to analyze the M. pellegrini population in the Longxi River, the results showed that this population had low genetic diversity, inbreeding, recent bottleneck and small effective population size because of isolation from the Changjiang River superadded human activities, it may face a threat if we do not take measures to protect the population.
6 DATA AVAILABILITY STATEMENTAll data generated and analyzed in this study are included in this manuscript.
Aljanabi S M, Martinez I. 1997. Universal and rapid saltextraction of high-quality genomic DNA for PCR-based techniques. Nucleic. Acids. Res., 25(22): 4 692-4 693.
DOI:10.1093/nar/25.22.4692 |
Allendorf F W, Luikart G H. 2007. Conservation and the Genetics of Populations. Blackwell Publishing, Malden, MA, USA; Oxford, UK; and Carlton, Victoria, Australia.
|
Bateman A J. 1948. lntra-sexual selection in Drosophila. Heredity, 2(3): 349-368.
DOI:10.1038/hdy.1948.21 |
Bendjilali N, Hsueh W C, He Q M, Willcox D C, Nievergelt C M, Donlon T A, Kwok P Y, Suzuki M, Willcox B J. 2014. Who are the Okinawans? Ancestry, genome diversity, and implications for the genetic study of human longevity from a geographically isolated population. J. Gerontol.: Ser. A, 69(12): 1 474-1 484.
DOI:10.1093/gerona/glt203 |
Bonnen P E, Pe'er I, Plenge R M, Salit J, Lowe J K, Shapero M H, Lifton R P, Breslow J L, Daly M J, Reich D E, Jones K W, Stoffel M, Altshuler D, Friedman J M. 2006. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat. Genetics, 38(2): 214-217.
DOI:10.1038/ng1712 |
Brook B W, Tonkyn D W, Q'Grady J J, Frankham R. 2002. Contribution of inbreeding to extinction risk in threatened species. Conserv. Ecol., 6(1): 16.
DOI:10.5751/ES-00387-060116 |
Chen Y Y. 1998. Fauna Sinica, Osteichthyes Cypriniformes Ⅱ. Science Press, Beijing.
(in Chinese)
|
Cornuet J M, Luikart G. 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144(4): 2 001-2 014.
|
Daniels S J, Priddy J A, Walters J R. 2000. Inbreeding in small populations of red-cockaded woodpeckers:insights from a spatially explicit individual-based model. Genetics, Demography and Viability of Fragmented Populations, 4: 129-147.
|
Do C, Waples R S, Peel D, Macbeth G M, Tillett B J, Ovenden J R. 2014. NeEstimator v2:re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour., 14(1): 209-214.
DOI:10.1111/men.2013.14.issue-1 |
Emlen S T, Oring L W. 1977. Ecology, sexual selection, and the evolution of mating systems. Science, 197(4300): 215-223.
DOI:10.1126/science.327542 |
Excoffier L, Lischer H E. 2010. Arlequin suite ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour., 10(3): 564-567.
DOI:10.1111/men.2010.10.issue-3 |
Frankham R, Ralls K. 1998. Conservation biology:inbreeding leads to extinction. Nature, 392(6675): 441-442.
DOI:10.1038/33022 |
Franklin I R. 1980. Evolutionary change in small populations. In: Soule M E, Wilcox B A eds. Conservation Biology— An Evolutionary-Ecological Perspective. Sinauer Associates, Sunderland, Massachusetts, USA. p.135-149. https://www.researchgate.net/publication/308155920_Evolutionary_Change_In_Small_Populations
|
Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN:two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics, 12(6): 543-548.
|
Gao X, Tan D Q, Liu H Z, Wang J W. 2009. Exploitation status and conservation of a population of Megalobrama pellegrini in Longxi River in the Upper Yangtze River basin. Sichuan J. Zool., 28(3): 329-333.
(in Chinese with English abstract) |
Gilpin M E. 1986. Minimum viable populations: processes of species extinction. In: Soulé M E ed. Conservation Biology: The Science of Scarcity and Diversity. Sinauer Associates, Sunderland, Massachusetts. p.19-34.
|
Grant W, Bowen B W. 1998. Shallow population histories in deep evolutionary lineages of marine fishes:insights from sardines and anchovies and lessons for conservation. J. Heredity, 89(5): 415-426.
DOI:10.1093/jhered/89.5.415 |
Grayson K L, Mitchell N J, Monks J M, Keall S N, Wilson J N, Nelson N J. 2014. Sex ratio bias and extinction risk in an isolated population of tuatara (Sphenodon punctatus). PLoS One, 9(4): e94214.
DOI:10.1371/journal.pone.0094214 |
Hauser L, Adcock G J, Smith P J, Ramírez J H B, Carvalho G R. 2002. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus). Proc. Natl. Acad. Sci. U.S.A., 99(18): 11 742-11 747.
DOI:10.1073/pnas.172242899 |
Huxley J. 1940. The New Systematics. Clarendon Press, Oxford. 596p.
|
Jones O R, Wang J L. 2010. COLONY:a program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour., 10(3): 551-555.
DOI:10.1111/men.2010.10.issue-3 |
Kalinowski S T, Wagner A P, Taper M L. 2006. ML-RELATE: a computer program for maximum likelihood estimation of relatedness and relationship. Mol. Ecol. Notes, 6(2): 576-579.
DOI:10.1111/men.2006.6.issue-2 |
Keller L F, Arcese P, Smith J N M, Hochachka W M, Stearns S C. 1994. Selection against inbred song sparrows during a natural population bottleneck. Nature, 372(6504): 356-357.
DOI:10.1038/372356a0 |
Klug H. 2011. Animal Mating Systems. In: eLS. John Wiley & Sons, Ltd., Chichester. https://doi.org/10.1002/9780470015902.a0022553.
|
Langella O. 2000. POPULATIONS 1.2: population genetic software, individuals or population distance, phylogenetic trees. http://bioinformatics.org/~tryphon/populations/.
|
Larkin M A, Blackshields G, Brown N P, Chenna R, McGettigan P A, McWilliam H, Valentin F, Wallace I M, Wilm A, Lopez R, Thompson J D, Gibson T J, Higgins D.G. 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23(21): 2 947-2 948.
DOI:10.1093/bioinformatics/btm404 |
Lee S L, Ng K K S, Saw L G, Lee C T, Muhammad N, Tani N, Tsumura Y, Koskela J. 2006. Linking the gaps between conservation research and conservation management of rare dipterocarps:a case study of Shorea lumutensis. Biol. Conserv., 131(1): 72-92.
DOI:10.1016/j.biocon.2006.02.005 |
Li W J, Wang J W, Xie C X, Tan D Q. 2007a. Age structure and growth characteristics of Megalobrama pellegrini-an endemic fish living only in upper reaches of Yangtze River. J. Fish. Sci. China, 14(2): 215-222.
(in Chinese with English abstract) |
Li W J, Wang J W, Xie C X, Tan D Q. 2007b. Reproductive biology and spawning habitats of Megalobrama pellegrini, an endemic fish in upper-reaches of Yangtze River basin. Acta Ecol. Sin., 27(5): 1 917-1 925.
(in Chinese with English abstract) |
Librado P, Rozas J. 2009. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1 451-1 452.
DOI:10.1093/bioinformatics/btp187 |
Lindenmayer D B, Peakall R. 2000. The Tumut experimentintegrating demographic and genetic studies to unravel fragmentation effects: a case study of the native Bush Rat. In: Young A G, Clarke G M eds. Genetics, Demography, and Viability of Fragmented Populations. Cambridge University Press, Cambridge, UK. p.173-201.
|
Luikart G, Allendorf F W, Cornuet J M, Sherwin W B. 1998. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Heredity, 89(3): 238-247.
DOI:10.1093/jhered/89.3.238 |
Luo Y L. 1990. A revision of fishes of the cyprinid genus Megalobrama. Acta Hydrobiol. Sin., 14(2): 160-165.
(in Chinese with English abstract) |
Madsen T, Stille B, Shine R. 1996. Inbreeding depression in an isolated population of adders Vipera berus. Biol. Conserv., 75(2): 113-118.
DOI:10.1016/0006-3207(95)00067-4 |
Mayr E. 1963. Animal Species and Evolution. Harvard University Press, Cambridge, Massachusetts. 797p.
|
Pardo L M, MacKay I, Oostra B, van Duijn C M, Aulchenko Y S. 2005. The effect of genetic drift in a young genetically isolated population. Ann. Human Genetics, 69(3): 288-295.
DOI:10.1046/J.1469-1809.2005.00162.x |
Service S, DeYoung J, Karayiorgou M, Roos J L, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha J A, Heutink P, Aulchenko Y, Oostra B, van Duijn C, Jarvelin M-R, Varilo T, Peddle L, Rahman P, Piras G, Monne M, Murray S, Galver L, Peltonen L, Sabatti C, Collins A, Freimer N. 2006. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat. Genetics, 38(5): 556-560.
DOI:10.1038/ng1770 |
Shriver M D, Jin L, Chakraborty R, Boerwinkle E. 1993. VNTR allele frequency distributions under the stepwise mutation model: a computer simulation approach. Genetics, 134(3): 983-993.
|
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol., 30(12): 2 725-2 729.
DOI:10.1093/molbev/mst197 |
Valdes A M, Slatkin M, Freimer N B. 1993. Allele frequencies at microsatellite loci:the stepwise mutation model revisited. Genetics, 133(3): 737-749.
|
van Oosterhout C, Hutchinson W F, Wills D P M, Shipley P. 2004. Micro-checker:software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes, 4(3): 535-538.
DOI:10.1111/men.2004.4.issue-3 |
Wang J J, Tong J G, Zhang Y G, Peng Z G. 2014. Study on the genetic diversity of two wild populations of Megalobrama pellegrini (Teleostei, Cyprinidae). Acta Hydrobiol. Sin., 38(5): 975-979.
(in Chinese with English abstract) |
Westemeier R L, Brawn J D, Simpson S A, Esker T L, Jansen R W, Walk J W, Kershner E L, Bouzat J L, Paige K N. 1998. Tracking the long-term decline and recovery of an isolated population. Science, 282(5394): 1 695-1 698.
DOI:10.1126/science.282.5394.1695 |
Xiao W H, Zhang Y P, Liu H Z. 2001. Molecular systematics of Xenocyprinae (Teleostei:Cyprinidae):taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol. Phylogenet. Evol., 18(2): 163-173.
DOI:10.1006/mpev.2000.0879 |
Yeh F C, Yang R C, Boyle T, Ye Z H, Mao J. 1997. PopGene Version 1.31, Microsoft Window-based Freeware for Population Genetic Analysis. Quick User Guide. Edmonton, Biology and Biotechnology Center, University of Alberta, Canada. https://sites.ualberta.ca/~fyeh/popgene.pdf.
|
 2019, Vol. 37
2019, Vol. 37