Institute of Oceanology, Chinese Academy of Sciences
Article Information
- ZHENG Li, TANG Wen-Qiao, ZHANG Ya, GUO Hongyi
- Comparative proteomic analysis of olfactory rosettes in anadromous Coilia nasus and resident Coilia nasus
- Journal of Oceanology and Limnology, 37(4): 1324-1332
- http://dx.doi.org/10.1007/s00343-019-8153-6
Article History
- Received May. 31, 2018
- accepted in principle Aug. 8, 2018
- accepted for publication Oct. 24, 2018
2 National Demonstration Center for Experimental Fisheries Science Education, Shanghai 201306, China;
3 Key Laboratory of Freshwater Fishery Germplasm Resources, Ministry of Agriculture, Shanghai Ocean University, Shanghai 201306, China
The Japanese grenadier anchovy (Coilia nasus) belongs to the family Engraulidae, order Clupeiformes, and is distributed widely in the coastal waters of China, e.g., the Changjiang (Yangtze) River and Huanghe (Yellow) River etc. (Jiang et al., 2012). Anadromous C. nasus individuals annually undergo the long-distance spawning migration from the coastal ocean up to fresh-water when the spawning period arrives. However, the resident population of C. nasus in lakes does not perform the behavior of long-distance spawning migration for unknown reasons (Zhu et al., 2014). Several studies in salmon and American eels have drawn the conclusion that functional olfactory ability is essential to perform accurate spawning migration (Døving et al., 1985; Yano and Nakamura, 1992; Barbin et al., 1998). The strong olfactory responses to natal stream water have also been found in lacustrine sockeye salmon (Oncorhynchus nerka) through blood oxygenation level-dependent functional magnetic resonance imaging (Bandoh et al., 2011). These studies indicated that olfaction may be of great importance for the spawning migration in fish.
At present, little is known regarding the olfactory signaling in C. nasus. However, relevant information can be acquired from other vertebrate species (Kaupp, 2010). The interaction of odorous ligands and specific receptors in olfactory neurons initiates a cascade of signal transduction (Mombaerts, 1999). Golf protein is able to stimulate olfactory-specific adenylyl cyclase (Pace et al., 1985) and then produced cAMP (Breer et al., 1990) opens a cyclic nucleotide-gated cation (CNG) channels (Nakamura and Gold, 1987) which can result in an influx of Na+ and Ca2+ and the cell depolarization. Moreover, Ca2+-activated chloride channels enable an efflux of Cl-, which facilitates further depolarization (Reisert et al., 2005; Nickell et al., 2007). The chemical signals are eventually converted into the electronic signal that is delivered to higher brain center. Termination of the olfactory response may occur at all steps of the pathway (Zhu et al., 2014). The G protein receptor kinase (GRK) that phosphorylates activated receptors contributes to olfactory desensitization (Peppel et al., 1997) and arrestins can desensitize the receptors (Mashukova et al., 2006). Regulator of G-protein signaling (RGS) apparently decreases the activity of adenylyl cyclase (Sinnarajah et al., 2001) and phosphodiesterase can hydrolyze cAMP, which promotes the termination of olfactory signaling. In addition, the removal of Ca2+ ions is modulated by Na+/Ca2+ exchange mechanisms, which contribute to mediating the rapid recovery of the odor response (Noé et al., 1997; Reisert and Matthews, 1998).
Some of the olfactory genes involved in the spawning migration have been demonstrated to be differentially expressed at the mRNA level in the different life stages of the wild anadromous Atlantic salmon while no differential expression of these genes was identified in non-anadromous Atlantic salmon (Johnstone et al., 2011). Previous studies have stated clearly that several olfactory receptor genes were significantly up-regulated at the mRNA levels in the olfactory rosettes of anadromous C. nasus compared to resident C. nasus during the breeding season (Zhu et al., 2016). However, mRNA expression may not correlate directly with the abundance of their protein products (Muers, 2011) and the mRNA levels might explain approximately 40% of the variability in protein levels (Schwanhäusser et al., 2011). And the key molecules involved in the spawning migratory behavior between anadromous C. nasus and resident C. nasus have still remained unknown. Thus, research carried on protein level is essential.
The transcriptomic database of C. nasus olfactory rosettes (accession number SRP100816) provided a good resource for analyzing the proteome. In the present work, we performed a comparative proteome analysis coupled with transcriptome analysis using isobaric tags for relative and absolute quantification (iTRAQ), which lays the foundation for further investigation of C. nasus spawning migration.
2 MATERIAL AND METHOD 2.1 Fishes and tissue samplesFemale anadromous C. nasus (body length 30± 2 cm) was collected from the Jingjiang section of the Changjiang River, Jiangsu Province, China at the end of April 2017 when they were migrating to spawning grounds (Fig. 1). The fish collection was performed with the help of fisherman ZHANG Huacai with the fishing license (No. Suchuanbu 2017 ZX-M019). Poyang Lake is an adjacent lake of Changjiang River and is divided into Hukou County, Xingzi County, Duchang County, Yongxiu County, and other areas. It is possible for anadromous C. nasus to enter to the Poyang Lake. Thus, female resident C. nasus (body length 22±2 cm) was collected from Poyang Lake in Duchang County, Jiangxi Province, China at the beginning of April 2017 before anadromous C. nasus has reached Poyang Lake to spawn. The fish collection was performed with the assistance of fisherman Baishan ZHAN holding the fishing license (No. 0400051). We chose the sampling sites with the expectation that the olfactory rosettes of migrating and pre-spawning C. nasus from Jingjiang would be more sensitive to the imprinted odors compared to Poyang Lake. The acquired live C. nasus were euthanized by an overdose of anesthetic (MS-222, Sigma-Aldrich, USA) in a separate plastic container contained with ice bags (-20 ℃). Before sampling, we made an incision in the abdomen and immediately checked the gonadal development phase. If the gonadal development phase was in phase Ⅲ, we collected the olfactory rosettes of C. nasus. The rinsed olfactory rosettes were placed into liquid nitrogen at once and subsequently delivered to the Shanghai Ocean University for further processing. The remains of the samples were preserved at Shanghai Ocean University for subsequent study. Samples from Jingjiang were labeled JJ, and the samples from Poyang Lake were labeled PY.
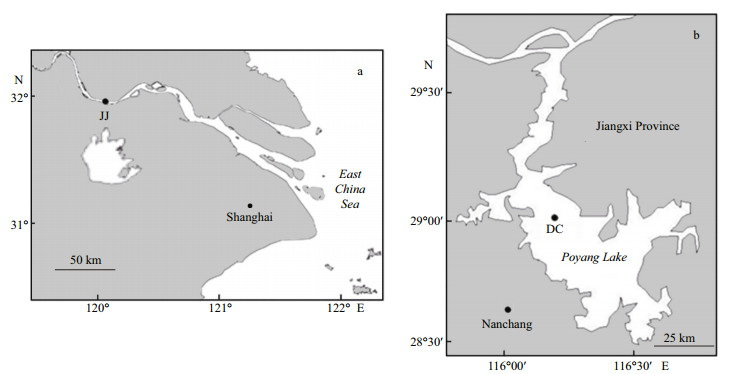
|
| Fig.1 Location of C. nasus sampling area JJ: the sampling site of Jingjiang section of the Changjiang River; DC: the sampling site of Poyang Lake in Duchang County. |
Olfactory rosettes from three similar individuals were pooled as one sample. The JJ group and the PY group respectively consisted of three biological replicates. Each sample was ground to a fine powder in liquid nitrogen and protein was extracted. The solution was centrifuged at 30 000×g for 15 min at 4 ℃. To the supernatant, 10 mmol/L DTT was added and the tube was incubated at 56 ℃ for 1 h. Subsequently, the tube was incubated for 1 h in the dark using 55 mmol/L iodoacetamide. After centrifugation, the precipitate was washed four times with chilled acetone for 2 h at -20 ℃ and was suspended in 0.5 mol/L tetraethylammonium bromide buffer. The protein concentration was detected using the Bradford method.
2.3 iTRAQ labeling and RP fractionationThe digested and desiccated peptides were labeled with iTRAQ 8-plex kits following the manufacturer's protocol. Three samples of the resident C. nasus were labeled with isobaric tags 113, 114, and 115, respectively. Three samples of the anadromous C. nasus were labeled with isobaric tags 116, 119, and 121, respectively. The peptides were separated on a high pH RP column. Elution was monitored by measuring absorbance at 214 nm.
2.4 LC-ESI-MS/MS analysis based on Triple TOF 5600 systemAll vacuum-dried fractions were resuspended in buffer A (2% ACN, 0.1% FA) and centrifuged at 20 000×g for 10 min. Subsequently, 10 μL of supernatant was loaded onto a C18 trap column attached to a Shimadzu LC-20AB HPLC Pump system. The fractions were analyzed using a Triple TOF 5600 system.
2.5 Protein identification and quantitative iTRAQ analysisThe MGF files converted from the raw MS/MS data were searched using Mascot (version 2.3.02) against the RNA-Seq transcriptomic database of C. nasus olfactory rosettes (accession number SRP100816). Quantitative analysis of the peptides was performed using the IQuant software. The results were filtered at 1% false discovery rate (FDR) at the peptide level. The proteins with a fold change of >1.2 and P-value less than 0.05 were recognized as the up-regulated proteins. The proteins with a fold change of < 0.83 fold change and P-value less than 0.05 were clustered as the down-regulated proteins.
These identified proteins were classified and grouped according to their Gene Ontology (GO) annotation, the Cluster of Orthologous Groups of proteins (COG) analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database. Differentially expressed proteins and proteins related to olfactory signaling were analyzed further. GO enrichment analysis and KEGG pathway enrichment analysis were performed to classify DEPs (differentially expressed proteins). Eventually, integrative analysis of the proteome and transcriptome was performed (the above work was completed with the help of BGI-Shenzhen).
3 RESULT 3.1 Protein identification and classificationUnder the 1% FDR threshold, a total of 5 408 proteins were identified on the basis of 40 729 unique spectra and 17 989 unique peptides. A total of 4 909 proteins (90.77%) were categorized with KEGG pathway and 320 pathways were identified in the Supplementary Information. Identified proteins analyzed with COGs were most represented by proteins in R (general function prediction only), J (translation, ribosomal structure and biogenesis) and O (posttranslational modification, protein turnover, chaperones). Moreover, a total of 3 560 proteins (65.83%) were functionally annotated with their GO. Numerous proteins were functioned in more than one of the GO terms.
3.2 Differentially expressed proteinsResident C. nasus was selected as the control group. Anadromous C. nasus was selected as the experimental group. We compared expressed proteins of olfactory rosettes in two groups. A total of 1 515 (629 up-regulated and 886 down-regulated) proteins exhibited expression differences with an FDR of less than 1%.
GO enrichment analysis was conducted to categorize DEPs (Supplementary Information). According to P-value, categories of up-regulated and down-regulated proteins are presented in Fig. 2. KEGG pathway enrichment analysis was performed to classify DEPs. According to P-value, categories of up-regulated and down-regulated proteins are shown in Fig. 3. KEGG pathway enrichment analysis of up-regulated proteins indicated statistically significant difference not only in olfactory transduction but also in the cGMP-PKG signaling pathway.
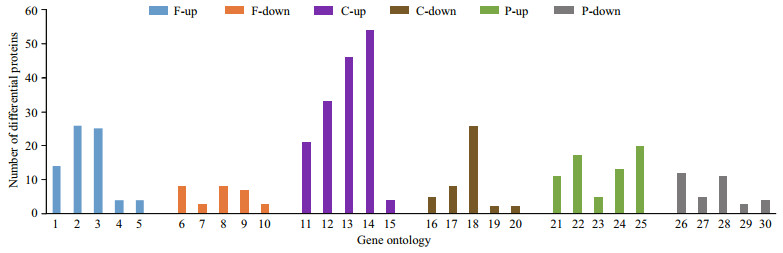
|
| Fig.2 GO enrichment analysis of the up-regulated and down-regulated proteins F: function; C: component; P: process. 1: iron ion binding; 2: substrate-specific transmembrane transporter activity; 3: ion transmembrane transporter activity; 4: oxygen transporter activity; 5: oxygen binding; 6: peptidase inhibitor activity; 7: cargo receptor activity; 8: peptidase regulator activity; 9: endopeptidase inhibitor activity; 10: scavenger receptor activity; 11: microtubule; 12: microtubule cytoskeleton; 13: cytoskeletal part; 14: cytoskeleton; 15: hemoglobin complex; 16: myosin filament; 17: myosin complex; 18: extracellular region; 19: protein phosphatase type 1 complex; 20: membrane attack complex; 21: protein polymerization; 22: microtubule-based movement; 23: gas transport; 24: cellular protein complex assembly; 25: microtubule-based process; 26: defense response; 27: defense response to bacterium; 28: immune response; 29: calcium ion-dependent exocytosis; 30: complement activation. |
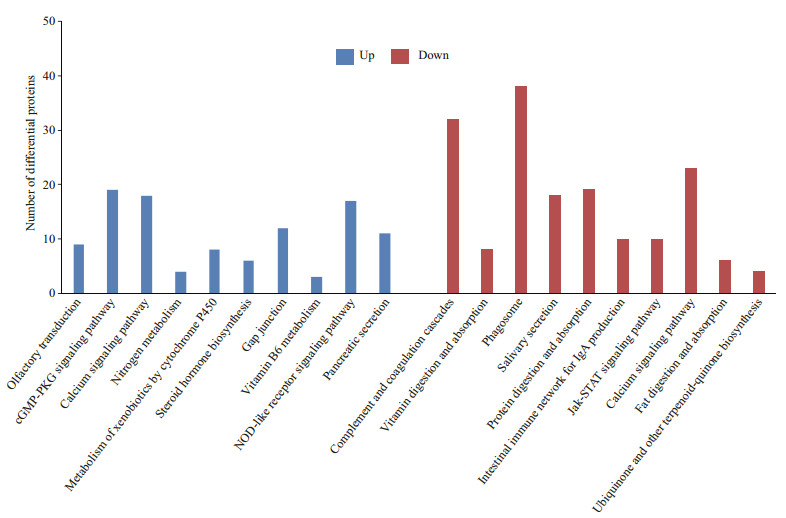
|
| Fig.3 KEGG pathway enrichment analysis of the up-regulated and down-regulated proteins |
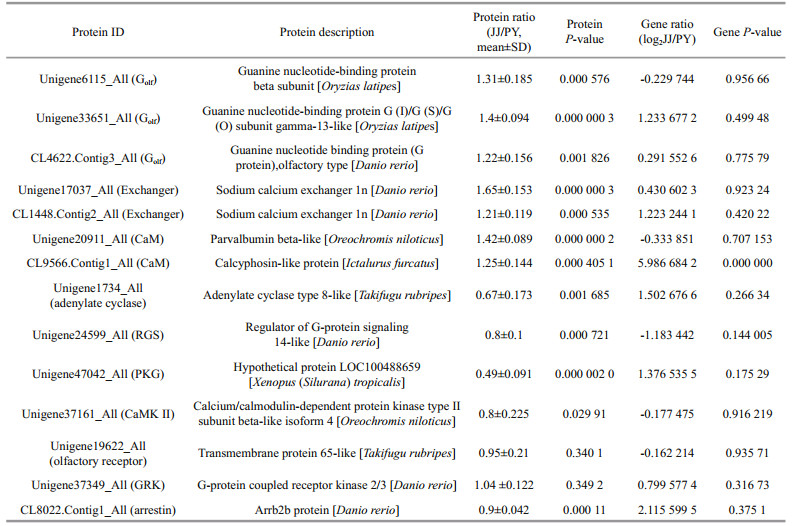
Especially, in view of the importance of olfactory signaling in spawning migration of anadromous C. nasus, we concentrated on DEPs involved in olfactory signaling. In our proteomic data, the expression of Golf protein and the sodium/calcium exchanger was found to be significantly up-regulated in the anadromous population compared with the resident population (Fig. 4). Our finding suggested a decrease in the expression of adenylate cyclase (Unigene1734_All) and RGS. The expression of cGMP-dependent protein kinase (PKG) was significantly down-regulated. The expression of Calmodulin (CaM) was increased and CaM-dependent protein kinase Ⅱ (CaMKⅡ) was decreased.
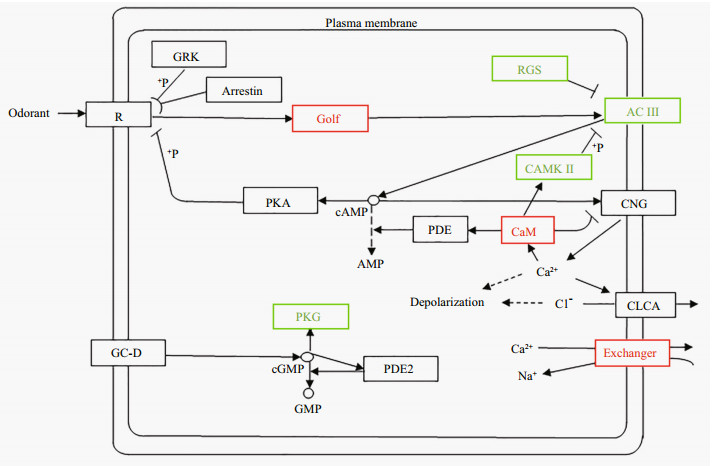
|
| Fig.4 Functional annotation of C. nasus proteins using the KEGG pathway of olfactory transduction Red: up-regulated; green: down-regulated. R: odorant receptor; Golf: G protein; AC: adenylate cyclase; CNG: cyclic nucleotide-gated cation channel; GC-D: guanylyl cyclase; CLCA: calcium-activated chloride channel; PDE: phosphodiesterase; PKG: cGMP-dependent protein kinase; RGS: regulator of G-protein signaling; PKA: cAMP-dependent protein kinase A; GRK: G protein receptor kinase; CAMKⅡ: calcium/calmodulin-dependent protein kinase Ⅱ; CAM: calmodulin. |
The quantitative analysis of the transcript was based on uploaded transcriptome (SRP100816) from our laboratory. It was possible to do the integrative analysis of transcriptome and proteome due to the same sampling strategies, including the same sampling sites and sampling season. One hundred fifty-five cases of concordant expression between DEGs (differentially expressed genes) and DEPs were evident (Supplementary Information). The number of cases of the opposite trend occurring in DEPs and DEGs was 69. A few genes (396) were only differentially expressed at the mRNA level and 1 276 significant changes were only observed at the protein level.
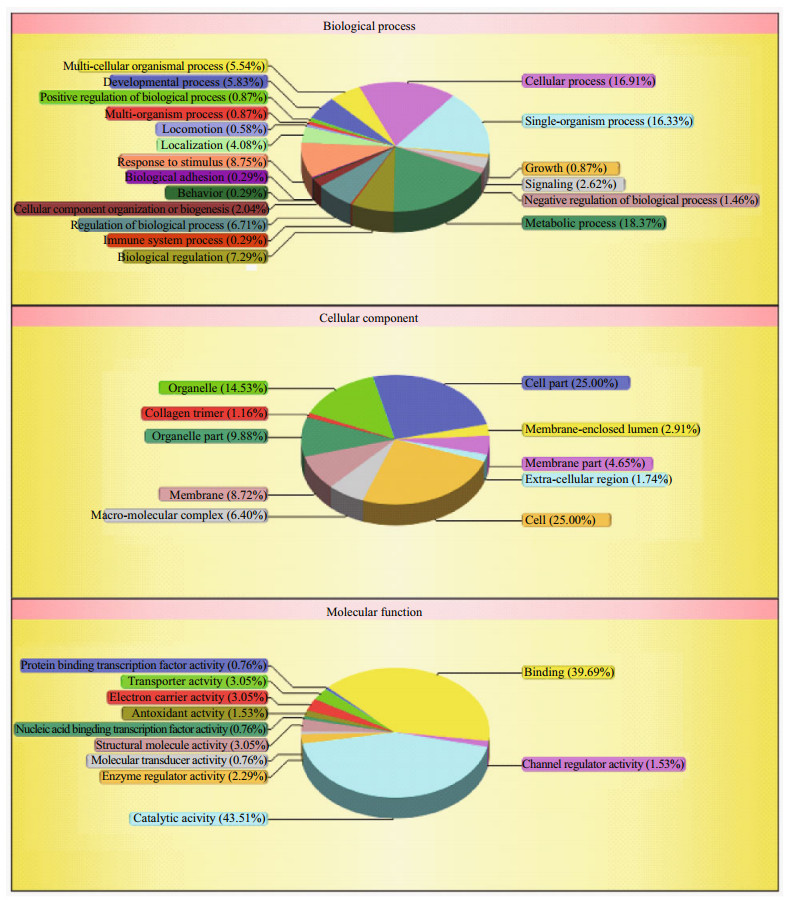
The enrichment analysis of GO and KEGG pathway were conducted to categorize these proteins having the same trend occurring in DEGs and DEPs (Supplementary Information). In the GO category of biological process, 18.37% of the proteins were involved in the metabolic process (Fig. 5). Of the proteins assigned to the GO category of cellular component, 25% of the proteins were involved in the cells. Additionally, of the proteins annotated with potential molecular function, catalytic activity (43.51%) and binding (39.69%) were enriched in this category. Among these KEGG pathway enrichment categories, the cluster for "metabolic pathways" was the largest, followed by "carbon metabolism" and "Glycolysis / Gluconeogenesis".
|
| Fig.5 The GO enrichment analysis of genes having the same trend occurring in DEGs and DEPs |
Although several identified proteins related to olfactory signaling were differentially expressed between the two populations at the protein level, they did not display significant changes at the mRNA level (Table 1). Proteins expression was not always consistent with gene expression.
|
Although the sampling time is different, the gonadal development phase of anadromous C. nasus and resident C. nasus was both in phase Ⅲ. In this study, we firstly seek to hunt for the proteomic changes in olfactory rosettes of two populations, especially changes involved in olfactory signaling. Generally, if C. nasus use specific genes to coordinate life stage-specific activities, then it is expected that these genes are expressed at higher levels. Compared with resident C. nasus, anadromous C. nasus may detect odorant cues in the water through their olfactory system, which may demand an increased expression level of some genes during the spawning migration. Previous relevant studies were through quantitative mRNA data to predict protein expression levels, which was indeed insufficient. Here, proteins expression related to olfactory signaling and some pathways were compared between resident C. nasus and anadromous C. nasus.
Vertebrate olfactory receptors are G protein-coupled receptors, which is crucial for a rapid and robust odorant recognition. Several distinct families of olfactory receptors include MORs (main olfactory receptors), V1Rs (vomeronasal type-1 receptors), V2Rs (vomeronasal type-2 receptors), TAARs (trace amine-associated receptors) and FPRs (formyl peptide receptors) (Zhu et al., 2016). And significant differences of some V1Rs at mRNA level were detected between the anadromous and resident C. nasus (Zhu et al., 2016). In our proteomic data, only an olfactory receptor (Unigene19622_All) was identified but displayed no expression difference. The up-regulated expression of Golf may indicate the important role in olfactory transduction during C. nasus spawning migration. The gene expression of the Golf did not display significant change between two populations at mRNA level. Expression changes at protein level may not be detected at the mRNA level at the same time (Hu et al., 2013). A series of regulatory processes play a vital role in controlling protein expression, which belongs to biological factors and may partially account for the complex relationship between mRNA and protein abundances (Vogel and Marcotte, 2012). Adenylyl cyclase and cAMP signaling are critical for olfactory-dependent behavior (Watt and Storm, 2001). Adenylate cyclase (Unigene1734_All) was unexpectedly downregulated in anadromous C. nasus compared with resident C. nasus. The expression of RGS was decreased, which may suggest a lower termination response and sustained detection to imprinted odors during spawning migration. These results indicate changes of proteins related to olfaction but do not indicate changes of the olfactory signaling pathway.
Previous studies have shown that salmon exposed to an odorant during a sensitive period for imprinting can show enhanced sensitivity of olfactory guanylyl cyclase activity to that odorant during their homeward migration (Dittman et al., 1997) and changes in olfactory-specific guanylyl cyclase increasing olfactory sensitization is important for natal stream recognition (Yamamoto et al., 2010). In addition, cGMP can activate adenylate cyclase in a cGMPdependent protein kinase (PKG) manner, generating a sustained cAMP signal upon odorant stimulation (Moon et al., 1998). KEGG pathway enrichment analysis of up-regulated proteins displayed the statistically significant difference in the cGMP-PKG signaling pathway. However, the expression of PKG (Unigene47042_All) in anadromous C. nasus relative to resident C. nasus was down-regulated at the protein level.
To the best of our knowledge, the Ras-MAPK (mitogen-activated protein kinase) pathway regulated the perception and transmission of sensory signals in olfactory neurons in Caenorhabditis elegans (Hirotsu et al., 2000). Furthermore, odorants can activate the MAPK pathway in rodent olfactory neurons due to CaMKⅡ phosphorylation (Watt and Storm, 2001). Thus, CaMKⅡ can link the termination of the cAMP signal by inhibiting adenylyl cyclase to activation of MAP kinase. The expression of CaMK Ⅱ (Unigene37161_All) was shown to be decreased.
The samples were from two different waters, which could have an impact on the results. Additionally, more studies are still needed to verify the described proteins and determine behavioral responses.
5 CONCLUSIONThis study exhibits a proteomic profile of olfactory rosettes of C. nasus and differently expressed proteins were detected between anadromous and resident C. nasus, which may provide a useful resource for research into the spawning migration of C. nasus and other molecular studies in C. nasus. Additionally, we speculate that Golf protein may play a crucial role in the olfactory signaling during the spawning migration of anadromous C. nasus.
6 DATA AVAILABILITY STATEMENTThe datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
7 ACKNOWLEDGMENTWe thank Mr. ZHAO Zhengguan and Dr. SONG Xiaojing for help for sample collection, and Dr. ZHU Guoli for completing transcriptome analysis of Japanese grenadier anchovy.
Electronic supplementary materialSupplementary material (Supplementary Information) is available in the online version of this article at https://doi.org/10.1007/s00343-019-8153-6.
Bandoh H, Kida I, Ueda H. 2011. Olfactory responses to natal stream water in sockeye salmon by BOLD fMRI. PLoS One, 6(1): e16051.
DOI:10.1371/journal.pone.0016051 |
Barbin G P, Parker S J, McCleave J D. 1998. Olfactory clues play a critical role in the estuarine migration of silver-phase American eels. Environmental Biology of Fishes, 53(3): 283-291.
DOI:10.1023/A:1007469232698 |
Breer H, Boekhoff I, Tareilus E. 1990. Rapid kinetics of second messenger formation in olfactory transduction. Nature, 345(6270): 65-68.
DOI:10.1038/345065a0 |
Dittman A H, Quinn T P, Nevitt G A, Hacker B, Storm D R. 1997. Sensitization of olfactory guanylyl cyclase to a specific imprinted odorant in coho salmon. Neuron, 19(2): 381-389.
DOI:10.1016/S0896-6273(00)80947-2 |
Døving K B, Westerberg H, Johnsen P B. 1985. Role of olfaction in the behavioral and neuronal responses of Atlantic salmon, Salmo salar, to hydrographic stratification. Canadian Journal of Fisheries and Aquatic Sciences, 42(10): 1 658-1 667.
DOI:10.1139/f85-207 |
Hirotsu T, Saeki S, Yamamoto M, Iino Y. 2000. The Ras-MAPK pathway is important for olfaction in Caenorhabditis elegans. Nature, 404(6775): 289-293.
DOI:10.1038/35005101 |
Hu G J, Koh J, Yoo M J, Grupp K, Chen S X, Wendel J F. 2013. Proteomic profiling of developing cotton fibers from wild and domesticated Gossypium barbadense. New Phytologist, 200(2): 570-582.
DOI:10.1111/nph.12381 |
Jiang T, Yang J, Liu H B, Shen X Q. 2012. Life history of Coilia nasus from the Yellow Sea inferred from otolith Sr:Ca ratios. Environmental Biology of Fishes, 95(4): 503-508.
DOI:10.1007/s10641-012-0066-6 |
Johnstone K A, Lubieniecki K P, Koop B F, Davidson W S. 2011. Expression of olfactory receptors in different life stages and life histories of wild Atlantic salmon (Salmo salar). Molecular Ecology, 20(19): 4 059-4 069.
DOI:10.1111/mec.2011.20.issue-19 |
Kaupp U B. 2010. Olfactory signalling in vertebrates and insects:differences and commonalities. Nature Reviews Neuroscience, 11(3): 188-200.
DOI:10.1038/nrn2789 |
Mashukova A, Spehr M, Hatt H, Neuhaus E M. 2006. β-arrestin2-mediated internalization of mammalian odorant receptors. The Journal of Neuroscience, 26(39): 9 902-9 912.
DOI:10.1523/JNEUROSCI.2897-06.2006 |
Mombaerts P. 1999. Molecular biology of odorant receptors in vertebrates. Annual Review of Neuroscience, 22: 487-509.
DOI:10.1146/annurev.neuro.22.1.487 |
Moon C, Jaberi P, Otto-Bruc A, Baehr W, Palczewski K, Ronnett G V. 1998. Calcium-sensitive particulate guanylyl cyclase as a modulator of cAMP in olfactory receptor neurons. The Journal of Neuroscience, 18(9): 3 195-3 205.
DOI:10.1523/JNEUROSCI.18-09-03195.1998 |
Muers M. 2011. Gene expression:transcriptome to proteome and back to genome. Nature Reviews Genetics, 12(8): 518.
DOI:10.1038/nrg3037 |
Nakamura T, Gold G H. 1987. A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature, 325(6103): 442-444.
DOI:10.1038/325442a0 |
Nickell W T, Kleene N K, Kleene S J. 2007. Mechanisms of neuronal chloride accumulation in intact mouse olfactory epithelium. The Journal of Physiology, 583: 1 005-1 020.
DOI:10.1113/jphysiol.2007.129601 |
Noé J, Tareilus E, Boekhoff I, Breer H. 1997. Sodium/calcium exchanger in rat olfactory neurons. Neurochemistry International, 30(6): 523-531.
DOI:10.1016/S0197-0186(96)00090-3 |
Pace U, Hanski E, Salomon Y, Lancet D. 1985. Odorant-sensitive adenylate cyclase may mediate olfactory reception. Nature, 316(6025): 255-258.
DOI:10.1038/316255a0 |
Peppel K, Boekhoff I, McDonald P, Breer H, Caron M G, Lefkowitz R J. 1997. G protein-coupled receptor kinase 3(GRK3) gene disruption leads to loss of odorant receptor desensitization. The Journal of Biological Chemistry, 272(41): 25 425-25 428.
DOI:10.1074/jbc.272.41.25425 |
Reisert J, Lai J, Yau K W, Bradley J. 2005. Mechanism of the excitatory Cl- response in mouse olfactory receptor neurons. Neuron, 45(4): 553-561.
DOI:10.1016/j.neuron.2005.01.012 |
Reisert J, Matthews H R. 1998. Na+-dependent Ca2+ extrusion governs response recovery in frog olfactory receptor cells. The Journal of General Physiology, 112(5): 529-535.
DOI:10.1085/jgp.112.5.529 |
Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature, 473(7347): 337-342.
DOI:10.1038/nature10098 |
Sinnarajah S, Dessauer C W, Srikumar D, Chen J, Yuen J, Yilma S, Dennis J C, Morrison E E, Vodyanoy V, Kehrl J H. 2001. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase Ⅲ. Nature, 409(6823): 1 051-1 055.
DOI:10.1038/35059104 |
Vogel C, Marcotte E M. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews Genetics, 13(4): 227-232.
DOI:10.1038/nrg3185 |
Watt W C, Storm D R. 2001. Odorants stimulate the ERK/mitogen-activated protein kinase pathway and activate cAMP-response element-mediated transcription in olfactory sensory neurons. The Journal of Biological Chemistry, 276(3): 2 047-2 052.
DOI:10.1074/jbc.M006703200 |
Yamamoto Y, Hino H, Ueda H. 2010. Olfactory imprinting of amino acids in lacustrine sockeye salmon. PLoS One, 5(1): e8633.
DOI:10.1371/journal.pone.0008633 |
Yano K, Nakamura A. 1992. Observations on the effect of visual and olfactory ablation on the swimming behavior of migrating adult chum salmon, Oncorhynchus keta. Japanese Journal of Ichthyology, 39(1): 67-83.
|
Zhu G L, Tang W Q, Wang L J, Wang C, Wang X M. 2016. Identification of a uniquely expanded V1R (ORA) gene family in the Japanese grenadier anchovy (Coilia nasus). Marine Biology, 163: 126.
DOI:10.1007/s00227-016-2896-9 |
Zhu G L, Wang L J, Tang W Q, Liu D, Yang J Q. 2014. De novo transcriptomes of olfactory epithelium reveal the genes and pathways for spawning migration in Japanese grenadier anchovy (Coilia nasus). PLoS One, 9(8): e103832.
DOI:10.1371/journal.pone.0103832 |
 2019, Vol. 37
2019, Vol. 37


