Institute of Oceanology, Chinese Academy of Sciences
Article Information
- ZHU Ying, LI Yangzhen, LI Hailong, WANG Lei, ZHANG Ning, LIU Yang, MENG Liang, XU Xiwen, DONG Zhongdian, WEI Min, GUO Hua, CUI Zhongkai, LI Xihong, SHAO Changwei, XU Wenteng
- iTRAQ-based analysis of 17β-estradiol induced proteome in Chinese tongue sole Cynoglossus semilaevis
- Journal of Oceanology and Limnology, 37(5): 1659-1668
- http://dx.doi.org/10.1007/s00343-019-8222-x
Article History
- Received Aug. 31, 2018
- accepted in principle Nov. 21, 2018
- accepted for publication Jan. 7, 2019
2 Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, China;
3 Research Institute of Metabolic Disease, Qingdao University, Qingdao 266003, China
Sexual dimorphism is particularly interesting in fish aquaculture because of its close association withfi sh growth and productivity. A number of fish species display sexual dimorphism, such as bigger males in Nile tilapia Oreochromis niloticus (Beardmore et al., 2001) and yellow catfish Pelteobagrus fulvidraco (Wang et al., 2009), and bigger females in flatfish like Bastard halibut Paralichthys olivaceus (Yoneda et al., 2007) and Chinese tongue sole Cynoglossus semilaevis (Chen et al., 2007). Thus, the productivity can be significantly enhanced by increasing the ratio of favored sex in specific fish species.
In North China, tongue sole is particularly favored for its taste and high meat quality. The female tongue sole is usually 2–4 times bigger than the male, so the increasing female ratio would benefit its aquatic yield. However, under existing aquaculture practices, male tongue sole is present in a higher proportion due to several reasons, e.g. genetic female sex-reversed to phenotypic male, which greatly limits the expansion of this industry. Improved understanding of sex determination and differentiation may facilitate biased production towards females.
Hormonal induction is widely used in producing monosex or sterile populations in aquaculture. As hormones operate on the developmental process via multiple regulatory pathways, unraveling the mechanism underpinning hormone induced sex changes could provide new perspectives for sex control. 17β-Estradiol (E2) is one of the most commonly used hormone to induce feminization and is effective in many fish species, including medaka Oryzias latipes (Kobayashi and Iwamatsu, 2005), Chinese tongue sole C. semilaevis (Zhang and Liu, 2009), bluegill sunfish Lepomis macrochirus (Chen et al., 2012), South American cichlid fish Cichlasoma dimerus (Zhang et al., 2011), Nile tilapia O. niloticus (Sun et al., 2015). In fish, E2 could exert its effect through several mechanisms: (Ⅰ) E2 activates estradiol receptors (ERs), which directly binds to estrogen response element (ERE) localized upstream of the target genes, resulting in transcriptional regulation (Nagler et al., 2010); (Ⅱ) ER interacts with other transcription factors (TFs), such as the luteinizing hormone β gene (Melamed et al., 2002), which then plays a role in gene regulation; (Ⅲ) E2 directly interacts with enzymes and activates the enzymatic functions, leading to rapid cascade reaction. For example, G protein-coupled receptors could involve in fish oocyte maturation in this way (Thomas, 2012). Although E2 receptors in fish have been well characterized (Hawkins et al., 2000; Ma et al., 2000; Choi and Habibi, 2003; Nagler et al., 2007), the thorough information of E2 regulatory circuit is still not clear. The OMIC analysis is especially advantageous in elucidating complex regulatory pathways. Recently, the proteomic approach was widely applied in fish to illustrate E2 regulatory network. Ibarz et al. (2013) used 2-D dimension electrophoresis to investigate the role of E2 on the skin and scale formation and regeneration in gilthead sea bream (Sparus aurata). Subsequently, DIGE was employed for screening proteins related to sexdimorphic behavior in brown ghost knifefish (Apteronotus leptorhynchus) after E2 treatment (Zupanc et al., 2014). Schilling et al. (2015) analyzed the protein expression patterns after E2 induction in white perch Morone americana using nanoLC-MS/ MS method.
OMIC studies was initiated in tongue sole since the deciphering of its genome in 2014 (Chen et al., 2014). These efforts lead to the understanding of transcriptomic changes across early developmental stages (Shao et al., 2017), epigenetic regulation of sex-reversal (Shao et al., 2014), and the characterization of sex determination/differentiation related genes (Li et al., 2016; Xu et al., 2016; Cui et al., 2017; Zhu et al., 2017). The progress has improved our knowledge of sex determination/differentiation process at genomic and transcriptomic levels. However, post-transcriptional regulation and modification may also play an important role in the biological process, thus a proteomic study will provide additional information by taking posttranscriptional and translational regulation into consideration and thoroughly illustrating the regulatory pathways.
In this study, we used the iTRAQ method to compare the E2 induced and uninduced proteomics aiming to (Ⅰ) obtain the proteomic overview of tongue sole and (Ⅱ) identify the key proteins in response to E2 induction, by which we try to enrich our knowledge of E2 regulatory network and shed light on sex control practice.
2 MATERIAL AND METHOD 2.1 Material17β-estradiol (E2) was purchased from a company (Solarbio, Beijing, China) and solubilized in ethanol to prepare the stock solution (300 mg/L). Tongue sole was obtained from Huanghai Base (Haiyang, Shandong, China). Immersion method was used and there are four experimental groups: control group, ethanol control group (1:10 000 dilution into water tank), E2 treated groups (concentrations were 10 and 30 μg/L, respectively). Each experimental group contained ~200 fish and was separately kept in 60 L water tank with an open circuit under 22–24℃. The treatment started at 30 days post hatching (dph) and ended at 90 dph. Fish were kept in the corresponding solution for 2 hours (8–10 a.m.) by switching off the water circuit and the remainder of the time in open circulating water.
2.2 Method 2.2.1 Ethics statementAll experiments were approved by the Animal Care and Use Committee, the Yellow Sea Fisheries Research Institute, China. Anesthetic was applied to reduce fish suffering during the experimental process.
2.2.2 Identification of genotypic and phenotypic sexForty fish were picked up from each group for sampling at 90 dph. Fins were cut and stored in ethanol for DNA isolation. As precise gonad cannot be obtained in tongue sole fry, the samples have mixed gonad, muscle, intestine and kidney, termed gonadal mixture in the following text. The samples were stored at -80℃ for isolation of RNA and protein, or fixed in Bouins solution for paraffin slice preparation and HE staining. Five fish were selected from each group (90 dph) for weight and length measurement. The genetic and phenotypic sex was determined according to the established methods (Chen et al., 2007, 2012).
2.2.3 Protein preparation, iTRAQ labeling, and LCMS/MS (liquid chromatography/tandem mass spectrometry) analysisAfter genotype identification by sex-specific primers, three males and three females from the control group and 30 μg/L estradiol group at 90 dph were picked. Protein was extracted by using a Minute TM Detergent-Free protein extraction kit separately (Invent Biotechnologies, MN, USA) and the concentration of protein was determined as previously described (Bradford, 1976). One milligram protein was picked up from each individual and mixed for further iTRAQ analysis, designated Ethanol Control (EC) and Estradiol 30 μg/L (E30). iTRAQ labeling and SCX fractionation were performed by Lianchuan Company (Lianchuan, Hanzhou, China).
2.2.4 Differential protein identification and bioinformatics analysisThe quantitative protein ratios were weighted and normalized by the median ratio in Mascot. Proteins with P < 0.05 and fold change over 1.2 were considered as significantly expressed. Gene Ontology (GO) was performed to classify the corresponding genes (http://www.geneontology.org) and there were three ontologies that can represent the molecular function, cellular component, and biological process, respectively. The protein structure was predicted using the SMART website (http://smart.emblheidelberg.de/).
2.2.5 qPCR and statistical analysisDifferentially expressed proteins (DEPs) identified by iTRAQ were selected for transcription evaluation by qPCR. Three males and three females were included in ethanol control and 30 μg/L E2 treated groups (90 dph). β-actin and rpl13α were used as internal references for normalization (Supplementary Table S1, Tang et al., 2007; Li et al., 2010). For each individual, the transcription level was evaluated using the comparative CT method and the experiments were performed in triplicates. A t-test was performed and the significant difference was only accepted when P < 0.05.
3 RESULT 3.1 Sex reversal of E2 inductionAmong the individuals whose phenotypic sex could be confirmed, all the genetic male individuals after E2 treatment showed sex-reversal (one from 10 μg/L and four from 30 μg/L) (Fig. 1), which supported the previous observation that E2 treatment led to an increased ratio of phenotypic female (Zhang and Liu, 2009). The E2 treatment had no consistent or significant effect on growth, when body length and weight were compared between E2 treated and control fish at 90 dph, and this is true for all experimental groups (control: average length 4.84±0.46 cm; average weight 0.50±0.14 g; ethanol control: average length 4.14±0.34 cm; average weight 0.40±0.07 g; 10 μg/L E2 treated: average length 4.84±0.39 cm; average weight 0.44±0.05 g; 30 μg/L E2 treated: average length 4.60±0.45 cm; average weight 0.52±0.11 g).

|
| Fig.1 Gonadal slides of genetic male (a–e) and female (f) tongue sole at 90 days post hatching (dph) a. genetic male from control group; b. genetic male from 10 μg/L estradiol treated group; c–e. genetic male from 30 μg/L estradiol treated group; f. genetic female form control group. SG: spermatogonium; OG: oogonium; OC: oocyte; OCA: ovarian cavity. |
The protein extract from three males and three females was subjected to SDS-PAGE. As shown in Supplementary Fig.S1a, no significant difference was observed between the EC and E30 groups. After iTRAQ analysis, 2 168 proteins were identified using the tongue sole genome data as a reference (Supplementary Fig.S1b, Supplementary Table S2). Based on the molecular weight, the proteins distributed between 20–60 kDa accounted for ~50% of the total protein. It was also worth noting that protein over 100 kDa accounted for about 20% of the total protein (Supplementary Fig.S1c). Using different GO annotation strategies, the proteins were clustered according to three categories: molecular function, cellular component or biological process, and the top 10 subcategories were listed (Fig. 2).

|
| Fig.2 GO annotation of the total protein obtained from the gonads of control and estradiol treated tongue sole |
Among the total 2 168 identified proteins, 409 proteins were identified as differentially expressed proteins (DEPs) after E2 treatment, including 259 upregulated and 150 downregulated ones. In the upregulated group, the relative proteins abundance of the E2-treated sample ranged from 1.20 to 6.31 fold of the control sample; In the down-regulated group, proteins abundance in the E2-treated sample were from 0.28 to 0.83 fold of the control sample (Supplementary Table S3).
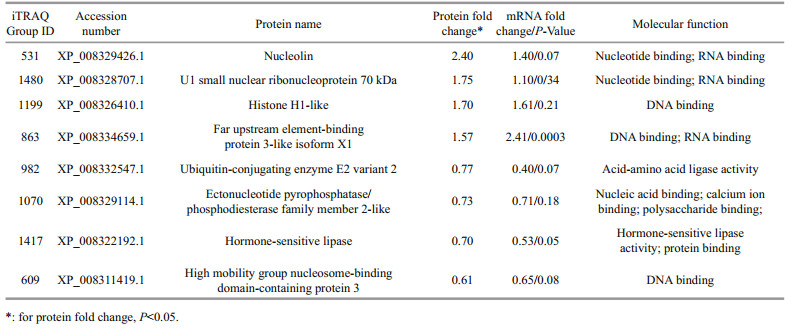
3.4 Transcriptional analysis of DEPsNineteen DEPs were further selected for transcriptional analysis after exclusion of the muscle related proteins. Eleven of the 19 genes were excluded from the study due to the low transcriptional abundance or large individual fluctuation. As shown in Table 1 and Fig. 3a, the other eight DEPs exhibited the same variation tendency in protein level. However, a significant difference was observed only in the far upstream element-binding protein 3-like isoform X1 (iTRAQ group ID, 863). To check whether the large fluctuation arises from a mixture of male and female samples, the transcription levels were further compared by separating male and female samples (Fig. 3b). We found some targets displayed the sex-biased transcription, such as female-biased expression of histone H1-like (1199). In general, the variation trends in male and female individuals after E2 induction are consistent in most targets except for nucleolin (531) and U1 small nuclear ribonucleoprotein (1480) that displayed increased expression in the female but vice versa in the male. When these targets were checked separately in male or female individuals displayed less individual variations (significant difference in targets 531, and 863) than the male ones (significant difference in target 863 only).

|
| Fig.3 Transcription analyses of eight DEPs in mixed samples (a) and different sex (b) Asterisk indicates significant difference (P < 0.05). |
The genome decoding and gene editing breakthrough promoted the study of sex determination and differentiation mechanism in tongue sole (Chen et al., 2014; Cui et al., 2017). However, most studies focused on genomic and transcription levels, the proteomic study was rare. In this work, 2 168 proteins were identified by iTRAQ analysis, accounting for ~10% of the total 21 516 annotated genes (Chen et al., 2014), and 409 were differentially expressed after E2 induction (Supplementary Table S2 and Supplementary Table S3, sheet 1). According to the typical motif 5′-GGTCAnnnTGACC-3′ of Estrogen Response Element (ERE) (Hall et al., 2001; Yaşar et al., 2016), seven candidates were found after screening the 409 DEPs (Supplementary Table S3, sheet 2). Despite all these motifs were located inside the coding sequences, the four and a half LIM domains proteins still attracted our interest because its motif was localized in the first exon, but whether this protein could act as an ERE protein requires further study.
A large part of the identified proteins was related to muscle composition. This may attribute to sampling process because the gonad of fry is not fully developed and the isolated samples were indeed a mixture of gonad and muscle. Therefore, we have re-selected the DEPs for further qPCR confirmation according to the following criteria: (1) exclusion of the muscle related proteins, such as actin, keratin. Although they might also respond to E2 induction, we focused on unraveling the gonadal proteomics in this study, so these muscle related proteins were not considered; (2) inclusion of more chromosomes. As in tongue sole, we have observed that gene related to sex determination and differentiation may accumulate in the same chromosome, for example, male-beneficial genes are enriched in the Z chromosome (Chen et al., 2014; Shao et al., 2014), and we propose these genes might exhibit the same tendency upon E2 treatment. Because of this, the inclusion of genes from different chromosomes would be better to avoid the probable 'chromosome-biased' effect and provide an all-sided overview to judge the effect of E2 treatment. Based on these rules, 19 DEPs were picked for transcription analysis (Supplementary Table S4).
Additional to the sampling limitation as indicated in the Materials and Methods section, the large standard deviation may be caused by sex-biased transcription. When the male and female samples were separated for analysis, the results were improved, e.g. 531 (Fig. 3). It is worth noting that the responses among female individuals seem to be more consistent than in the male. Indeed, a similar phenomenon exists during the vaccination process in human, where sexbiased responses were observed (Klein et al., 2015). However, this phenomenon needs to be confirmed in further study, and we might perform androgen treatment and compare the responses between different sexes.
Zhang and Liu (2009) proposed that E2 treatment could lead to sex-reversal of tongue sole by comparing phenotypic sex ratio among different groups, while the genetic sex was not determined due to lack of molecular marker. Using the same experimental procedure and combining genetic sex identification method, we have confirmed that E2 treatment can per se induce the feminization of tongue sole in this study. Among the DEPs with top change fold, we found the proteins that were previously reported to involve in E2 induction process (Nagler et al., 2010), e.g. vitellogenin and vitellogenin-2 with 6.3 and 3.6 times boost after E2 treatment, although their mRNA levels exhibited large individual deviation (Supplementary Table S4). This individual deviation of VTG is also observed in other fish, such as fathead minnows, European flounder, and zebra finch, either under a natural condition or upon E2 treatment (George, 2004; Han et al., 2009; Feswick et al., 2017).
In order to identify new partners during E2 induction network, the function of the eight DEPs was analyzed from bioinformatics level. These DEPs include two enzymes, amino acid ligase, and hormonesensitive lipase, and six proteins involved either in DNA binding or RNA binding (Table 1). Regarding the versatile regulatory strategy of E2 in teleost (reviewed in Pinto et al., 2014), it is not surprising that these proteins were identified. Based on the previous reports, we classified the DEPs into three groups, e.g. E2 responsive, a specific function (development, sex differentiation, immunity and so on), and general function.
For E2 responsive elements, it is reported that hormone-sensitive lipase can respond to multiple hormones in human and fish, including E2, insulin, and growth hormone (Palin et al., 2003; Khieokhajonkhet et al., 2016; Sun et al., 2017). Another interesting finding is that the histone H1-like protein could be up-regulated by E2 treatment in tongue sole. In fact, a similar study was conducted in fathead minnows (Pimephales promelas), where the treatment of 17α-ethinylestradiol resulted in downregulation of four histone proteins. The E2 influence on histone level suggests a possible link between hormone and epigenetic regulation (Martyniuk et al., 2010).
Ubiquitin-conjugating enzyme E2 variant 2 is a key enzyme in ubiquitination pathway and its involvement in spermatogenesis was reported in shrimp, fish, and mouse (Baarends et al., 1999; Leelatanawit et al., 2009; Hu and Chen, 2013). In this study, the downregulation of this enzyme after E2 treatment seemed to further support that it was a 'male-beneficial element'. For ectonucleotide pyrophosphatase/phosphodiesterase, it functions during embryonic and neural development and widely reported to various human disease (reviewed by Cholia et al., 2015), and a recent study in zebrafish also supported its important role in early embryogenesis (Frisca et al., 2016). It is worthy of further investigation why E2 treatment could affect this 'early stage' protein, which might provide clues for its new functional exploitation. For U1 small nuclear ribonucleoprotein 70 kDa (snRNP70), it is a component of the nuclear ribonucleoprotein complex, which acts on splicing of precursor mRNA. The cleavage of snRNP70 was proposed to facilitate the apoptosis process (Casciola et al., 1994; Tewari et al., 1995). As gonadal cell after E2 treatment did exhibit some apoptosis-like patterns (data not shown), we postulate that snRNP70 might be a promising candidate to link E2 and apoptosis.
Other DEPs seemed to function at a more general or basic level, including nucleolin, high mobility group nucleosome-binding domain-containing protein 3 (HMGN3), and far upstream elementbinding protein. Nucleolin is a multifunctional protein participating in ribosome biogenesis, chromatin organization, DNA/RNA RNA metabolism (Jia et al., 2017). HMG proteins represent a superfamily that could bind to DNA and nucleosomes. They play important roles in chromatin confirmation, DNA repair, transcription and loss of function is closely associated with numerous disease (West et al., 2001; Hock et al., 2007). As the only candidate significantly up-regulated (1.57 fold) by E2, far upstream elementbinding protein also increased to 1.79 fold after E2 treatment in human microvascular endothelium (TIME) cell line (Felty, 2010). However, this protein also participates in many biological processes, including transcription, mRNA stability, and translation (Zhang and Chen, 2012). Thus, the detailed roles of these three proteins during E2 induction is to be determined by further investigation.
5 CONCLUSIONin this study, we found e2 treatment could induce sex-reversal of genetic male tongue sole to phenotypic female. the itraq-based analysis was performed and 409 differentially expressed proteins (deps) were identified after e2 induction, including 259 upregulated and 150 down-regulated proteins. furthermore, among 19 randomly selected deps, eight of them exhibited a similar transcript tendency and the functions of the eight proteins included mainly the nucleotide binding. interestingly, a far upstream element-binding protein 3-like isoform displayed significant upregulation at both transcription and translation levels upon e2 induction. this work would facilitate our understanding of e2 regulatory mechanism from protein level.
6 DADA AVAILABILITY STATEMENTAll data generated and/or analyzed during this study are included in this published article and its supplementary information files.
7 ACKNOWLEDGEMENTWe would like to thank Professor Deborah M Power for the critical comments and language polishing.
Electronic supplementary materialSupplementary material (Supplementary Fig.S1 and Supplementary Tables S1–S4) is available in the online version of this article at https://doi.org/10.1007/s00343-019-8222-x.
Baarends W M, Hoogerbrugge J W, Roest H P, Ooms M, Vreeburg J, Hoeijmakers J H J, Grootegoed J A. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Developmental Biology, 207(2): 322-333.
DOI:10.1006/dbio.1998.9155 |
Beardmore J A, Mair G C, Lewis R I. 2001. Monosex male production in finfish as exemplified by tilapia:applications, problems, and prospects. Aquaculture, 197(1-4): 283-301.
DOI:10.1016/S0044-8486(01)00590-7 |
Bradford M M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72(1-2): 248-254.
DOI:10.1016/0003-2697(76)90527-3 |
Casciola-Rosen L A, Miller D K, Anhalt G J, Rosen A. 1994. Specific cleavage of the 70-kDa protein component of the U1 small nuclear ribonucleoprotein is a characteristic biochemical feature of apoptotic cell death. The Journal of Biological Chemistry, 269(49): 30757-30760.
|
Chen S L, Ji X S, Shao C W, Li W L, Yang J F, Liang Z, Liao X L, Xu G B, Xu Y, Song W T. 2012. Induction of mitogynogenetic diploids and identification of WW superfemale using sex-specific SSR markers in half-smooth tongue sole (Cynoglossus semilaevis). Marine Biotechnology, 14(1): 120-128.
DOI:10.1007/s10126-011-9395-2 |
Chen S L, Li J, Deng S P, Tian Y S, Wang Q Y, Zhuang Z M, Sha Z X, Xu J Y. 2007. Isolation of female-specific AFLP markers and molecular identification of genetic sex in half-smooth tongue sole (Cynoglossus semilaevis). Marine Biotechnology, 9(2): 273-280.
DOI:10.1007/s10126-006-6081-x |
Chen S L, Zhang G J, Shao C W, Huang Q F, Liu G, Zhang P, Song W T, An N, Chalopin D, Volff J N, Hong Y H, Li Q Y, Sha Z X, Zhou H L, Xie M S, Yu Q L, Liu Y, Xiang H, Wang N, Wu K, Yang C G, Zhou Q, Liao X L, Yang L F, Hu Q M, Zhang J L, Meng L, Jin L J, Tian Y S, Lian J M, Yang J F, Miao G D, Liu S S, Liang Z, Yan F, Li Y Z, Sun B, Zhang H, Zhang J, Zhu Y, Du M, Zhao Y F, Schartl M, Tang Q S, Wang J. 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nature Genetics, 46(3): 253-260.
DOI:10.1038/ng.2890 |
Choi C Y, Habibi H R. 2003. Molecular cloning of estrogen receptor α and expression pattern of estrogen receptor subtypes in male and female goldfish. Molecular and Cellular Endocrinology, 204(1-2): 169-177.
DOI:10.1016/S0303-7207(02)00182-X |
Cholia R P, Nayyar H, Kumar R, Mantha A K. 2015. Understanding the multifaceted role of ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2) and its altered behaviour in human diseases. Current Molecular Medicine, 15(10): 932-943.
DOI:10.2174/1566524015666150921104804 |
Cui Z K, Liu Y, Wang W W, Wang Q, Zhang N, Lin F, Wang N, Shao C W, Dong Z D, Li Y Z, Yang Y M, Hu M Z, Li H L, Gao F T, Wei Z F, Meng L, Liu Y, Wei M, Zhu Y, Guo H, Cheng C H K, Schartl M, Chen S L. 2017. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Scientific Reports, 7: 42213.
DOI:10.1038/srep42213 |
Felty Q. 2011. Proteomic 2D DIGE profiling of human vascular endothelial cells exposed to environmentally relevant concentration of endocrine disruptor PCB153 and physiological concentration of 17β-estradiol. Cell Biology and Toxicology, 27(1): 49-68.
DOI:10.1007/s10565-010-9170-6 |
Feswick A, Isaacs M, Biales A, Flick R W, Bencic D C, Wang R L, Vulpe C, Brown-Augustine M, Loguinov A, Falciani F, Antczak P, Herbert J, Brown L, Denslow N D, Kroll K J, Lavelle C, Dang V, Escalon L, Garcia-Reyero N, Martyniuk C J, Munkittrick K R. 2017. How consistent are we? Interlaboratory comparison study in fathead minnows using the model estrogen 17α-ethinylestradiol to develop recommendations for environmental transcriptomics. Environmental Toxicology and Chemistry, 36(10): 2614-2623.
DOI:10.1002/etc.3799 |
Frisca F, Colquhoun D, Goldshmit Y, Änkö M L, Pébay A, Kaslin J. 2016. Role of ectonucleotide pyrophosphatase/phosphodiesterase 2 in the midline axis formation of zebrafish. Scientific Reports, 6: 37678.
DOI:10.1038/srep37678 |
George S, Gubbins M, MacIntosh A, Reynolds W, Sabine V, Scott A, Thain J. 2004. A comparison of pollutant biomarker responses with transcriptional responses in European flounders (Platicthys flesus) subjected to estuarine pollution. Marine Environmental Research, 58(2-5): 571-575.
DOI:10.1016/j.marenvres.2004.03.047 |
Hall J M, Couse J F, Korach K S. 2001. The multifaceted mechanisms of estradiol and estrogen receptor signaling. The Journal of Biological Chemistry, 276(40): 36869-36872.
DOI:10.1074/jbc.R100029200 |
Han D, Haunerland N H, Williams T D. 2009. Variation in yolk precursor receptor mRNA expression is a key determinant of reproductive phenotype in the zebra finch (Taeniopygia guttata). Journal of Experimental Biology, 212(9): 1277-1283.
DOI:10.1242/jeb.026906 |
Hawkins M B, Thornton J W, Crews D, Skipper J K, Dotte A, Thomas P. 2000. Identification of a third distinct estrogen receptor and reclassification of estrogen receptors in teleosts. Proceedings of the National Academy of Sciences of the United States of America, 97(20): 10751-10756.
DOI:10.1073/pnas.97.20.10751 |
Hock R, Furusawa T, Ueda T, Bustin M. 2007. HMG chromosomal proteins in development and disease. Trends in Cell Biology, 17(2): 72-79.
DOI:10.1016/j.tcb.2006.12.001 |
Hu Q M, Chen S L. 2013. Cloning, genomic structure and expression analysis of ubc9 in the course of development in the half-smooth tongue sole (Cynoglossus semilaevis). Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 165(3): 181-188.
DOI:10.1016/j.cbpb.2013.03.007 |
Ibarz A, Pinto P I S, Power D M. 2013. Proteomic approach to skin regeneration in a marine teleost:modulation by oestradiol-17β. Marine Biotechnology (NY), 15(6): 629-646.
DOI:10.1007/s10126-013-9513-4 |
Jia W Y, Yao Z Y, Zhao J J, Guan Q B, Gao L. 2017. New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sciences, 186: 1-10.
DOI:10.1016/j.lfs.2017.07.025 |
Khieokhajonkhet A, Kaneko G, Hirano Y, Wang L, Ushio H. 2016. Different effects of growth hormone and fasting on the induction patterns of two hormone-sensitive lipase genes in red seabream Pagrus major. General and Comparative Endocrinology, 236: 121-130.
DOI:10.1016/j.ygcen.2016.06.025 |
Klein S L, Marriott I, Fish E N. 2015. Sex-based differences in immune function and responses to vaccination. Transactions of the Royal Society of Tropical Medicine and Hygiene, 109(1): 9-15.
DOI:10.1093/trstmh/tru167 |
Kobayashi H, Iwamatsu T. 2005. Sex reversal in the medaka Oryzias latipes by brief exposure of early embryos to estradiol-17β. Zoological Science, 22(10): 1163-1167.
DOI:10.2108/zsj.22.1163 |
Leelatanawit R, Sittikankeaw K, Yocawibun P, Klinbunga S, Roytrakul S, Aoki T, Hirono I, Menasveta P. 2009. Identification, characterization and expression of sexrelated genes in testes of the giant tiger shrimp Penaeus monodon. Comparative Biochemistry and Physiology Part A:Molecular & Integrative Physiology, 152(1): 66-76.
DOI:10.1016/j.cbpa.2008.09.004 |
Li H L, Xu W T, Zhang N, Shao C W, Zhu Y, Dong Z D, Wang N, Jia X D, Xu H, Chen S L. 2016. Two Figla homologues have disparate functions during sex differentiation in halfsmooth tongue sole (Cynoglossus semilaevis). Scientific Reports, 6: 28219.
DOI:10.1038/srep28219 |
Li Z J, Yang L J, Wang J, Shi W C, Pawar R A, Liu Y M, Xu C G, Cong W H, Hu Q R, Lu T Y, Xia F, Guo W, Zhao M, Zhang Y Y. 2010. β-Actin is a useful internal control for tissue-specific gene expression studies using quantitative real-time PCR in the half-smooth tongue sole Cynoglossus semilaevis challenged with LPS or Vibrio anguillarum. Fish & Shellfish Immunology, 29(1): 89-93.
DOI:10.1016/j.fsi.2010.02.021 |
Ma C H, Dong K W, Yu K L. 2000. cDNA cloning and expression of a novel estrogen receptor β-subtype in goldfish (Carassius auratus). Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression, 1490(1-2): 145-152.
DOI:10.1016/S0167-4781(99)00235-3 |
Martyniuk C J, Kroll K J, Doperalski N J, Barber D S, Denslow N D. 2010. Environmentally relevant exposure to 17α-ethinylestradiol affects the telencephalic proteome of male fathead minnows. Aquatic Toxicology, 98(4): 344-353.
DOI:10.1016/j.aquatox.2010.03.007 |
Melamed P, Koh M, Preklathan P, Bei L, Hew C. 2002. Multiple mechanisms for Pitx-1 transactivation of a luteinizing hormone β subunit gene. The Journal of Biological Chemistry, 277: 26200-26207.
DOI:10.1074/jbc.M201605200 |
Nagler J J, Cavileer T, Sullivan J, Cyr D G, Rexroad Ⅲ C. 2007. The complete nuclear estrogen receptor family in the rainbow trout:discovery of the novel ERα2 and both ERβ isoforms. Gene, 392(1-2): 164-173.
DOI:10.1016/j.gene.2006.12.030 |
Nagler J J, Davis T L, Modi N, Vijayan M M, Schultz I. 2010. Intracellular, not membrane, estrogen receptors control vitellogenin synthesis in the rainbow trout. General and Comparative Endocrinology, 167(1): 326-330.
DOI:10.1016/j.ygcen.2010.03.022 |
Palin S L, McTernan P G, Anderson L A, Sturdee D W, Barnett A H, Kumar S. 2003. 17β-estradiol and anti-estrogen ICI:compound 182, 780 regulate expression of lipoprotein lipase and hormone-sensitive lipase in isolated subcutaneous abdominal adipocytes. Metabolism, 52(4): 383-388.
DOI:10.1053/meta.2003.50088 |
Pinto P I S, Estêvão M D, Power D M. 2014. Effects of estrogens and estrogenic disrupting compounds on fish mineralized tissues. Marine Drugs, 12(8): 4474-4494.
DOI:10.3390/md12084474 |
Schilling J, Nepomuceno A I, Planchart A, Yoder J A, Kelly R M, Muddiman D C, Daniels H V, Hiramatsu N, Reading B J. 2015. Machine learning reveals sex-specific 17β-estradiol-responsive expression patterns in white perch (Morone americana) plasma proteins. Proteomics, 15(15): 2678-2690.
DOI:10.1002/pmic.201400606 |
Shao C W, Bao B L, Xie Z Y, Chen X Y, Li B, Jia X D, Yao Q L, Ortí G, Li W H, Li X H, Hamre K, Xu J, Wang L, Chen F Y, Tian Y S, Schreiber A M, Wang N, Wei F, Zhang J L, Dong Z D, Gao L, Gai J W, Sakamoto T, Mo S D, Chen W J, Shi Q, Li H, Xiu Y J, Li Y Z, Xu W T, Shi Z Y, Zhang G J, Power D M, Wang Q Y, Schartl M, Chen S L. 2017. The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nature Genetics, 49(1): 119-124.
DOI:10.1038/ng.3732 |
Shao C W, Li Q Y, Chen S L, Zhang P, Lian J M, Hu Q M, Sun B, Jin L J, Liu S S, Wang Z J, Zhao H M, Jin Z H, Liang Z, Li Y Z, Zheng Q M, Zhang Y, Wang J, Zhang G J. 2014. Epigenetic modification and inheritance in sexual reversal of fish. Genome Research, 24(4): 604-615.
DOI:10.1101/gr.162172.113 |
Sun A, Wang T Z, Wang N, Liu X F, Sha Z X, Chen S L. 2015. Establishment and characterization of an ovarian cell line from half-smooth tongue sole Cynoglossus semilaevis. Journal of Fish Biology, 86(1): 46-59.
DOI:10.1111/jfb.12535 |
Sun J, Yang Z, Xiao P Z, Liu Y, Ji H, Du Z Y, Chen L Q. 2017. Two isoforms of hormone-sensitive lipase b are generated by alternative exons usage and transcriptional regulation by insulin in grass carp (Ctenopharyngodon idella). Fish Physiology and Biochemistry, 43(2): 539-547.
DOI:10.1007/s10695-016-0308-1 |
Tang R Y, Dodd A, Lai D, McNabb W C, Love D R. 2007. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochimica et Biophysica Sinica, 39(5): 384-390.
DOI:10.1111/j.1745-7270.2007.00283.x |
Tewari M, Beidler D R, Dixit V M. 1995. CrmA-inhibitable cleavage of the 70-kDa protein component of the U1 small nuclear ribonucleoprotein during Fas-and tumor necrosis factor-induced apoptosis. The Journal of Biological Chemistry, 270(32): 18738-18741.
DOI:10.1074/jbc.270.32.18738 |
Thomas P. 2012. Rapid steroid hormone actions initiated at the cell surface and the receptors that mediate them with an emphasis on recent progress in fish models. General and Comparative Endocrinology, 175(3): 367-383.
DOI:10.1016/j.ygcen.2011.11.032 |
Wang D, Mao H L, Chen H X, Liu H Q, Gui J F. 2009. Isolation of Y-and X-linked SCAR markers in yellow catfish and application in the production of all-male populations. Animal Genetics, 40(6): 978-981.
DOI:10.1111/j.1365-2052.2009.01941.x |
West K L, Ito Y, Birger Y, Postnikov Y, Shirakawa H, Bustin M. 2001. HMGN3a and HMGN3b, two protein isoforms with a tissue-specific expression pattern, expand the cellular repertoire of nucleosome-binding proteins. The Journal of Biological Chemistry, 276(28): 25959-25969.
DOI:10.1074/jbc.M101692200 |
Xu W T, Li H L, Dong Z D, Cui Z K, Zhang N, Meng L, Zhu Y, Liu Y, Li Y Z, Guo H, Ma J L, Wei Z F, Zhang N W, Yang Y M, Chen S L. 2016. Ubiquitin ligase gene neurl3 plays a role in spermatogenesis of half-smooth tongue sole (Cynoglossus semilaevis) by regulating testis protein ubiquitination. Gene, 592(1): 215-220.
DOI:10.1016/j.gene.2016.07.062 |
Yaşar P, Ayaz G, Muyan M. 2016. Estradiol-Estrogen receptor α mediates the expression of the CXXC5 gene through the estrogen response element-dependent signaling pathway. Scientific Reports, 6: 37808.
DOI:10.1038/srep37808 |
Yoneda M, Kurita Y, Kitagawa D, Ito M, Tomiyama T, Goto T, Takahashi K. 2007. Age validation and growth variability of Japanese flounder Paralichthys olivaceus off the Pacific coast of northern Japan. Fisheries Science, 73(3): 585-592.
DOI:10.1111/j.1444-2906.2007.01371.x |
Zhang B, Wang X L, Sha Z X, Yang C G, Liu S S, Wang N, Chen S L. 2011. Establishment and characterization of a testicular cell line from the half-smooth tongue sole, Cynoglossus semilaevis. International Journal of Biological Sciences, 7(4): 452-459.
DOI:10.7150/ijbs.7.452 |
Zhang J, Chen Q M. 2012. Far upstream element binding protein 1:a commander of transcription, translation and beyond. Oncogene, 32(24): 2907-2916.
DOI:10.1038/onc.2012.350 |
Zhang X Y, Liu H J. 2009. Effects of 17β-Estradiol on sex differentiation and growth in half-smooth tongue-sole(Cynoglossus semilaevis). Journal of Northeast Agricultural University, 40(6): 67-72.
(in Chinese with English abstract) |
Zhu Y, Hu Q M, Xu W T, Li H L, Guo H, Meng L, Wei M, Lu S, Shao C W, Wang N, Yang G P, Chen S L. 2017. Identification and analysis of the β-catenin1 gene in halfsmooth tongue sole (Cynoglossus semilaevis). PLoS One, 12(5): e0176122.
DOI:10.1371/journal.pone.0176122 |
Zupanc G K H, Ilieş I, Sîrbulescu R F, Zupanc M M. 2014. Large-scale identification of proteins involved in the development of a sexually dimorphic behavior. Journal of Neurophysiology, 111(8): 1646-1654.
DOI:10.1152/jn.00750.2013 |
 2019, Vol. 37
2019, Vol. 37