Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LI Wenjie, CHENG Jiao, HUI Min, SHA Zhongli
- Molecular phylogeny of the genus Clibanarius (Decapoda: Anomura: Diogenidae) based on mitochondrial and nuclear DNA sequences
- Journal of Oceanology and Limnology, 37(5): 1686-1697
- http://dx.doi.org/10.1007/s00343-019-8329-0
Article History
- Received Nov. 15, 2018
- accepted in principle Dec. 24, 2018
- accepted for publication Feb. 20, 2019
2 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China;
3 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, China;
4 University of Chinese Academy of Sciences, Beijing 100049, China
Hermit crabs are anomurans decapod crustaceans of the superfamily Paguroidea, which included more than 1 100 species (de Grave et al., 2009; McLaughlin et al., 2010). Most of the hermit crabs have an asymmetrical abdomen secluding in a shell of a gastropod. Although hermit crabs have colonized a range of inhabits from terrestrial to the deep sea (Tsang et al., 2011), they are best known from intertidal areas. As active carrion scavengers, hermit crabs are ecologically important groups of zoobenthos. (Williams and McDermott, 2004). Hermit crabs also represent an intermediate group of crustaceans from Macrura to Brachyura, which occupys an important position in crustacean evolution (Sha et al., 2015). Diogenidae Ortmann, 1892 is the second largest family of the superfamily Paguroidea. Most species of Diogenidae live in the ocean, and only one species has been reported from freshwater so far (See McLaughlin and Murray, 1990). Clibanarius Dana, 1852 is one of the speciose genera of Diogenidae and usually inhabits intertidal zones. It currently contains 59 species and 43 of them are known from the Indo-West Pacific (McLaughlin et al., 2007, 2010; Malay et al., 2018). They are commonly found in tropical or subtropical shallow water inhabiting sandy, estuary, mangroves, seagrass beds, rocky shore, and coral reef (Hirose et al., 2010; Sha et al., 2015). Most species of Clibanarius are small and share a superficial resemblance in morphology, increasing the difficulty of identification. The color patterns in the life of most Clibanarius species is often used as a diagnostic character for the species identification (Sha et al., 2015). It has been reported, however, that color differences within samples may not represent the different species (Knowlton and Mills, 1992; Hirose et al., 2010). In addition, the body color of hermit crabs easily fades away in alcohol, which makes identification more difficult.
Taxonomic groupings of Clibanarius have been proposed by Asakura (2005) based on the relative length of dactyls and propodi. According to Asakura's proposal, the species of Clibanarius can be divided into two morphological groups: (ⅰ) species have the dactyls shorter than the propodi of the second and third pereopods; (ⅱ) species have the dactyls equal to or slightly longer than the propodi of the second and third pereopods (Asakura, 2005). However, this taxonomic division has not been verified based on molecular data. In contrast to the wealth information on the phylogenetic relationships among the major taxa of Anomura (e.g., Ahyong et al., 2009; Tsang et al., 2011; Bracken-Grissom et al., 2013), few studies have focused on the interspecific relationships within Clibanarius (Hirose et al., 2010; Negri et al., 2014; Yoshikawa et al., 2018). Hirose et al. (2010) performed a preliminary phylogenetic analysis of Clibanarius based on partial sequences of the mitochondrial cytochrome c oxidase subunit Ⅰ (COI) gene, in which poorly resolved phylogenies prevented phylogenetic relationships of Clibanarius from an in-depth investigation. The close association of C. humilis, C. merguiensis and C. englaucus and the close relationships of C. longitarsus and C. striolatus were strongly supported. Negri et al. (2014) undertook phylogenetic analyses of selected Clibanarius species using the barcode region of the COI gene to distinguish C. symmetricus from C. vittatus. However, most nodes of the phylogenetic tree showed low support values. They only showed the close relationships between C. symmetricus, C. vittatus, C. lineatus, and C. sclopetarius. Yoshikawa et al. (2018) identified three specimens as Clibanarius virescens that had unknown coloration for Clibanarius based on the mitochondrial COI and nuclear histone H3 genes, in which phylogenetic relationships of Clibanarius species had received little attention. Overall, the interspecific relationship of Clibanarius is remained poorly resolved and the hypothesis of Asakura (2005) remained no attention.
It should be noted that the above-mentioned molecular phylogenies are based on analysis of a limited amount of sequence data, mainly mitochondrial DNA sequences, which could not provide enough phylogenetic resolutions. Two nuclear protein-coding genes, sodium-potassium ATPase α-subunit (NaK) and phosphoenolpyruvate carboxykinase (PEPCK), play elementary roles in multitudinous life forms and maintain good conservation throughout evolution (Tsang et al., 2008). These two genes have been widely used to recovery the phylogenetic relationships among higher taxonomic category in insects for many years (Friedlander et al., 1996; Wiegmann et al., 2000; Leys et al., 2002; Danforth et al., 2004, 2011). Recently, these two nuclear protein-coding genes have been increasingly utilized to resolve the phylogeny of the caridean shrimps (Li et al., 2011) and Anomura (Tsang et al., 2011). The mitochondrial DNA reveals speedy substitution saturation that restricts their practicability in resolving deep nodes, whereas the nuclear proteincoding genes are easy to align and provide a rich source of information that promotes a more precise recovery of phylogeny (Tsang et al., 2008). Therefore, the combination of mitochondrial and nuclear genes which has successfully resolved some controversial phylogenetic relationships is expected to provide more accurate phylogenetic information (Kiwaidae, Roterman et al., 2018).
To investigated phylogenetic relationships of 11 Indo-West Pacific Clibanarius species, both mitochondrial (16S rRNA and COI) and nuclear (NaK and PEPCK) genes are sequenced. The present study aims to examine whether phylogenetic groupings based on molecular data support or refute previously proposed taxonomic divisions and to elucidate the interspecific relationships within Clibanarius. Furthermore, we also aim to compare the practicability of mitochondrial genes or nuclear protein-coding genes in exploring interspecific relationships of Clibanarius. Establishing phylogenetic relationships of Clibanarius species will improve the understanding of taxonomic classification and evolutionary relationships of Anomura.
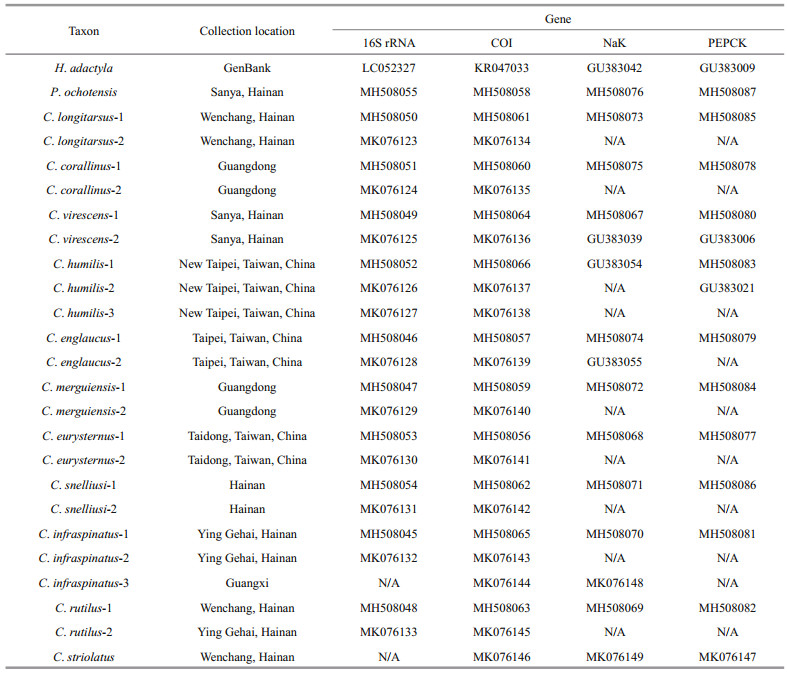
2 METERIAL AND METHOD 2.1 Sample collectionA total of 11 representatives of Clibanarius Dana, 1852 are utilized in this study, (detailed sampling information shown in Table 1). Species of the specimens are identified morphologically according to McLaughlin et al. (2007) and Sha et al. (2015). Among them, Clibanarius corallinus (Milne Edwards, 1848), C. virescens (Krauss, 1843), C. merguiensis de Man, 1888, C. englaucus Ball and Haig, 1972 and C. rutilus Rahayu, 1999 have ambulatory dactyl shorter than the propodus, while C. longitarsus (de Haan, 1833-1850), C. striolatus Dana, 1852, C. infraspinatus (Hilgendorf, 1869), C. eurysternus (Hilgendorf, 1879), C. humilis (Dana, 1851), and C. snelliusi Buitendijk, 1937 have ambulatory dactyl equal to or slightly longer than the propodus. Specimens used in this study are preserved in the Marine Biological Museum (MBM) in the Institute of Oceanology, Chinese Academy of Sciences, Qingdao (IOCAS). The outgroup taxa, namely Pagurus ochotensis Brandt, 1851 and Hippa adactyla (Fabricius, 1787), are chosen to root the phylogenetic trees. All the samples are preserved in 75%–95% ethanol.
|
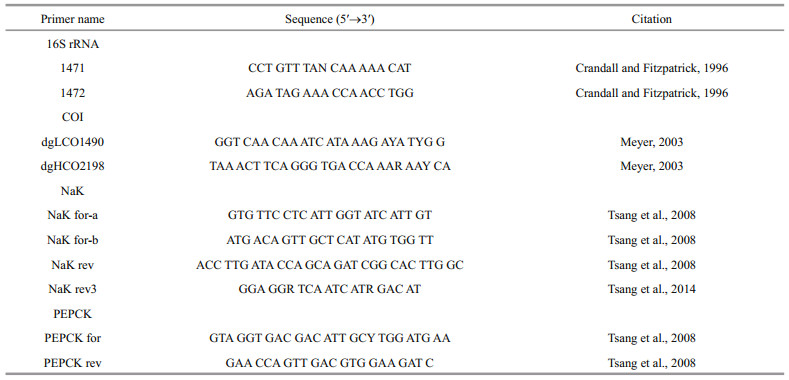
The pereopod or cheliped of each hermit crab (about 15 mg) is used for DNA extraction using a QIAGEN DNeasy Blood and Tissue Kit. The total genomic DNA is eluted in 200 μL of sterile distilled H2O and stored in -20℃ freezer. Partial fragments from four genes, 16S rRNA (~500 bp), COI (~660 bp), NaK (~590 bp), and PEPCK (~590 bp), are amplified by polymerase chain reaction (PCR). The amplifications are executed in 25 μL volumes that incorporated 1–2 μL of template DNA, 1 μL forward and reverse primers, PCR Mix containing 1 μmol/L RED Taq, 22 mmol/L Tris-HCL, 55 mmol/L KCl, 1.65 mmol/L MgCl2, and 220 μmol/L dNTP. The primers used are revealed in Table 2. For 16S rRNA and COI gene segments, the program cycling conditions are as follows: initial denaturation for 3 min at 94℃, followed by 33 cycles of 30 s at 94℃, 30 s at 50℃, and 50 s at 72℃, with a final extension at 72℃ for 3 min. The following primer combinations are used to amplify NaK and PEPCK gene segments: NaK for-a/NaK rev3, NaK for-b/NaK rev3, NaK for-b/NaK rev and PEPCK for/PEPCK rev, respectively (Tsang et al., 2008, 2014). The PCR profile for NaK is as follows: 3 min at 94℃ for initial denaturation, then 33 cycles of 30 s at 94℃, 30 s at 50–55℃ depending on individual samples, 50 s at 72℃ with a final extension at 72℃ for 3 min. The same program cycling conditions are employed for PEPCK with the extended time of 1 min 30 s at 72℃. The PCR products are purified by using of QIAquick gel purification kit (Qiagen) before sequencing. Purified PCR products are bidirectionally sequenced with ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) using the same primer pairs for PCR amplification. The sequences of the four genes of Hippa adactyla are obtained from GenBank. Similarly, NaK sequences of C. virescens-2, C. humilis-1, C. englaucus-2 and PEPCK sequences of C. virescens-2 and C. humilis-2 are acquired from GenBank. All sequences generated in the present study have been accessioned in GenBank, and corresponding accession numbers (MH508045– MH508087, MK076123–MK076149) are given in Table 1.
The chromatograms of forward and reverse sequence fragments are checked and assembled using SeqMan in the DNASTAR Lasergene software package. The inferred amino acid sequences are obtained to detect the presence of nonsense mutation using the program MEGA 6.0 (Tamura et al., 2013). Molecular diversity indices, such as the number of polymorphic sites, parsimony information sites, the insertions or deletions of nucleotide sequences, are achieved by Arlequin 3.5 (Excoffier and Lischer, 2010). PAUP*4.0b10 (Swofford, 2002) is used to examine the base compositional homogeneity within the taxa implemented by the Chi-square test. The sequences of the four genes are aligned separately using MUSCLE default settings that integrated into MEGA 6.0. After alignment, we splice the nucleotide sequences of the four/two genes to form a combined dataset for downstream phylogenetic analyses.
The phylogenetic trees are reconstructed using Maximum likelihood (ML) and Bayesian inference (BI) algorithms. The ML analysis is accomplished on the ATGC bioinformatics platform with PhyML 3.0 (Guindon et al., 2010), and Bayesian inference (BI) is conducted using MrBayes 3.2 (Huelsenbeck and Ronquist, 2001). In total, we analyze three datasets: all four concatenated genes, two concatenated mitochondrial genes, and two concatenated nuclear genes. The best-fit models of nucleotide substitution of each gene are selected using jModelTest v2.1.6 (Darriba et al., 2012). The best-fit model of the four concatenated genes and the concatenated two mitochondrial genes is GTR+I+G. The optional model of the concatenated two nuclear genes is TIM2+I+G. The best-fit models shown by the jModelTest for the 16S rRNA, COI, and NaK, PEPCK are TIM3+I+G, GTR+I+G, TIM1+G, and TIM2ef+G, respectively.
We conduct the ML analysis using the best-fit models of DNA substitution for the combined mitochondrial genes, nuclear genes, and all the four genes. For each analysis, if the model selected by the jModelTest is not available in the MrBayes and PhyML 3.0, we would use the most similar model to instead. The supporting value for each clade is estimated by 1 000 bootstrap replicates (BP). In a hybrid model, the partitioning of sequence data that come from multiple genes can provide a better estimation of phylogenetic relationships (Castoe et al., 2004; Brandley et al., 2005; Brown and Lemmon, 2007). Thus, we undertake the partitioned Bayesian analyses for each concatenated dataset, allowing specific substitution models to be assigned to each gene. A metropolis coupled Monte Carlo Markov Chains (MCMCMC) sampling is performed with four chains running simultaneous for 10 000 000 generations with trees sampled every 1 000 generations. The average standard deviations of the split frequencies (< 0.01) and potential scale reduction factor (RSRF, close to 1.0) are used as the criterions to validate the convergence of the analysis. After discarding the first 25% of the generations as burn-in, the 50% majority-rule consensus trees from the remaining trees are constructed to estimate posterior probabilities (PP) for each clade.
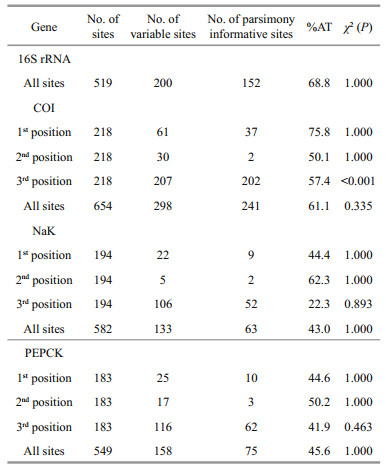
3 RESULT 3.1 Sequence characteristicsThe combined dataset of the four gene fragments consists of 2 304 nucleotide sites. The 16S rRNA and COI sequences show a strong AT bias, with mean overall values of 68.8% and 61.1%, respectively. Amongst the 519 sites of 16S rRNA, 200 are variable and 152 are parsimony informative. Amongst the 654 sites in COI, 298 characters are variable, 241 are parsimony informative. Sequences of PEPCK are slightly G/C biased (54.4%), whilst the NaK is also slightly G/C biased (57.0%). The aligned sequences of NaK gene includes 582 nucleotide positions with 133 variable sites and 63 parsimony informative sites. The PEPCK gene includes 549 positions in which 158 are variable and 75 are parsimony informative. No stop codons or indel are observed from the COI, NaK, and PEPCK. A total of 31-bp deletion is detected in the 16S rRNA sequence alignment. There is no significant base heterogeneity across all codon positions in the three protein-coding genes (Chisquare P>0.05), except in the third position of the COI gene (Table 3).
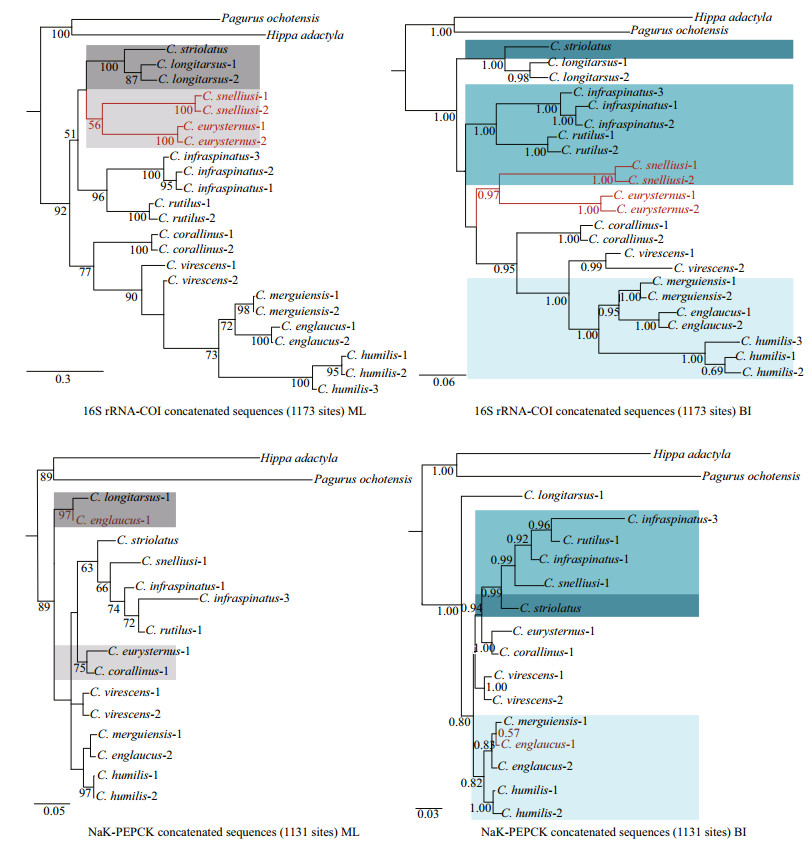
Unexpectedly, a discrepancy between the results of mitochondrial and nuclear DNA datasets is found in the present study (Fig. 1). Moreover, the two analytic methods (ML and BI) yield different results for each concatenated dataset. The difference between the tree topologies received by ML and BI methods of the mitochondrial concatenated dataset is the position of the species C. snelliusi and C. eurysternus. These two species cluster together and form a clade with C. striolatus, C. longitarsus, C. infraspinatus and C. rutilus using the ML method (BP=51%), while form a clade with C. corallinus, C. virescens, C. merguiensis, C. englaucus, and C. humilis using the BI method (PP < 0.50). The difference between the tree topologies received by ML and BI methods of the nuclear concatenated dataset is the position of C. englaucus-1. C. englaucus-1 and C. longitarsus occupy the basal position in the ML tree (BP < 50%), while C. englaucus-1 cluster with C. merguiensis-1, C. englaucus-2, and C. humilis in the BI tree (PP=0.82). Nevertheless, all the analyses reveal consistently the close relationship among C. merguiensis, C. englaucus, and C. humilis. On the other hand, the tree topologies of the four-gene concatenated dataset are congruent regardless of the methods analyzed. Here, only the BI tree is presented with support values for both ML and BI analyses (Fig. 2). The interspecific phylogenetic relationships of Clibanarius are better resolved by using all four genes concatenated dataset, which could not be clearly uncovered in the other two datasets with lower support values. For these reasons, we use this consensus tree as the phylogenetic tree of Clibanarius.
|
| Fig.1 Phylogenetic relationships of Clibanarius species based on concatenated mitochondrial genes (16S rRNA and COI) and concatenated nuclear genes (NaK and PEPCK) separately The ML and BI trees constructed from mitochondrial DNA and nuclear DNA datasets were shown. The bootstrap replicated of ML tree >50 and the posterior probabilities of BI tree >0.50 are noted. A discrepancy between the results of mitochondrial and nuclear DNA datasets by using the same analysis are marked by different background colors. A discrepancy between the results of two analytic methods by using the same dataset is marked by different font colors. |
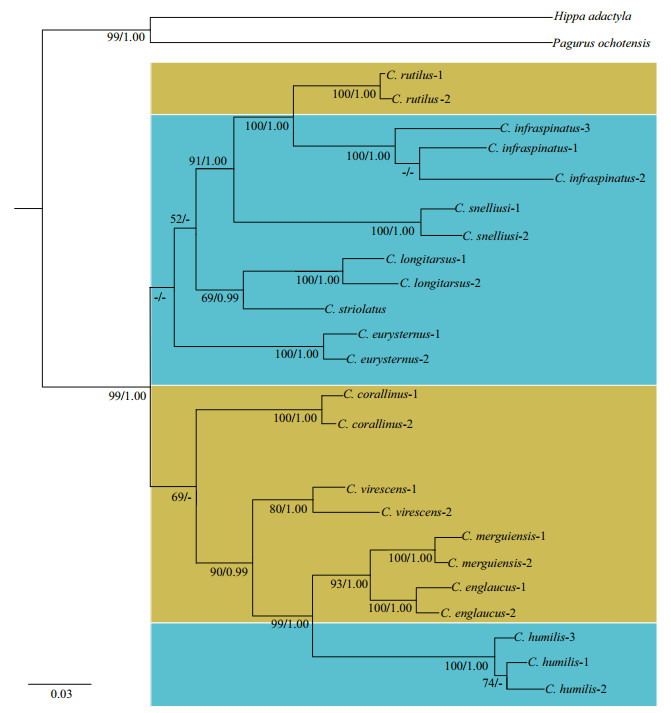
|
| Fig.2 Phylogenetic relationships of Clibanarius species based on the all four-gene concatenated dataset The BI topology was presented with support values for both ML and BI analyses. Nodal support values from left to right represent maximum likelihood bootstrap replicates (BP) / Bayesian posterior probabilities (PP), respectively. Species having the dactyls shorter than the propodi in the second and third pereopods are marked by the yellow background color. Species having the dactyls equal to or slightly longer than the propodi in the second and third pereopods are marked by the blue background color. The nodal support values of ML tree less than 50 and the posterior probabilities less than 0.95 are indicated by "-" |
As shown in Fig. 2, the tree topologies derived from the ML and BI analyses are highly congruent and some deep nodes are strongly supported. The clustering of C. rutilus with the dactyl shorter than the propodus and C. infraspinatus with the dactyl longer than the propodus is strongly supported by the analyses (BP=100%, PP=1.00) and form a clade with C. snelliusi with high support values (BP=91%, PP=1.00). C. longitarsus and C. striolatus cluster together with relatively high support values (BP=69%, PP=0.99), while the phylogenetic trees show relatively low support values for their relationship with C. rutilus, C. infraspinatus and C. snelliusi (BP=52%, PP=0.90). The branching positions of C. eurysternus and C. corallinus are not obtained in the current tree topologies. C. virescens represents an early divergence from C. merguiensis, C. englaucus and C. humilis with high support values (BP=90%, PP=0.99). Therein, C. merguiensis and C. englaucus with the dactyl shorter than the propodus clustered as a sister taxa with high supports (BP=93%, PP=1.00), forming a clade with C. humilis with the dactyl equal or subequal to the propodus (BP=99%, PP=1.00). It is worth noting that the supports values of some nodes for the interspecific relationships in the molecular tree are relatively low and further studies with more taxa sampling and genetic information are required to resolve this issue.
4 DISCUSSION 4.1 Implication of comparative studies on ClibanariusThis is the first study in which we investigated the phylogenetic relationship of Clibanarius based on both mitochondrial and nuclear DNA sequences. Previous studies on Clibanarius or other genera of diogenid phylogeny were based on morphological characteristics or/and few molecular data that relied on a single mitochondrial gene (Mantelatto et al., 2006; Tirelli et al., 2008, 2010; Hirose et al., 2010; Negri et al., 2014). Therein, they all showed lower support values and discrepancies in the critical nodes of Clibanarius phylogeny. In this study, we have explored the potential impact of the amount of sequence as well as the inclusion of multiple genes with distinct evolutionary histories on the phylogenetic reconstructions of Clibanarius. The lower resolution of interspecific phylogenetic relationships of Clibanarius is observed when mitochondrial DNA and nuclear DNA dataset are analyzed separately. Therein, two analyses methods product different tree topologies by using same dataset. By contrast, most of the interspecific relationships are robust and coincident in both ML and BI analyses based on the combined mitochondrial and nuclear DNA dataset. It has been recommended that long DNA sequences can yield a sufficient number of characters for robust phylogenetic inference and provide high phylogenetic resolution of relationships among closely related congeneric species (Minegishi et al., 2005; Cheng et al., 2011). The relatively short DNA sequences used for phylogeny construction may provide an explanation for the inconsistent phylogenetic patterns observed in previous reconstructions of Clibanarius phylogeny (Hirose et al., 2010; Negri et al., 2014).
In this study, we also demonstrate the importance of including different gene regions of molecular markers in the phylogenetic resolution of relationships among Clibanarius species. The characteristics of high copy number, rapid evolution rate and maternally inherited of mitochondrial genes makes them widely used as a molecular marker in phylogenetic studies (Moritz et al., 1987; Toon et al., 2009). Nevertheless, the rapid rate of nucleotide substitution among mitochondrial genes has limited their utility in higher levels of divergence (Toon et al., 2009). On the contrary, some of the nuclear protein-coding genes have the advantage of lower nucleotide substitution rate and are easy to align, which exhibit better performance in resolving phylogenetic relationships at deeper taxonomic levels (Moriyama and Powell, 1997; Chu et al., 2009). Therefore, a multigene phylogenetic approach is needed to construct a phylogeny, with slower-evolving genes settling deeper relationships and faster-evolving genes settling closer relationships (Palero and Crandall, 2009). It has been reported that a well-resolved topology is obtained to reconstruct phylogenetic relationships of the infraorder Stenopodidea (Chen et al., 2016) using mitochondrial and nuclear DNA sequences from a combined perspective. We consider it more likely that different mechanisms of nucleotide substitution and different evolutionary properties may explain the discrepancy between mitochondrial and nuclear DNA phylogenies of Clibanarius and suggest the significance of considering different gene regions of molecular markers in future phylogenetic considerations of Clibanarius.
4.2 Phylogenetic relationshipsAsakura (2005) proposed that members of Clibanarius could be separated into two morphological groups according to the relative length of dactyls and propodi. One group contained species having stout second and third pereopods with the dactyls shorter than the propodi. The other group was composed of species with the dactyls as long as or slightly longer than the propodi, resulting in slender second and third pereopods. However, the phylogenetic results herein contradicts Asakura (2005) morphological classification scheme for Clibanarius. C. rutilus with the dactyl shorter than the propodus forms a sister relationship to C. infraspinatus with the dactyl longer than the propodus. The same pattern is found in the close relationships with C. merguiensis, C. englaucus and C. humilis, in which C. merguiensis and C. englaucus with the dactyl shorter than the propodus cluster as a sister taxon, forming a clade with C. humilis with the dactyl equal to the propodus. Overall, the present molecular data indicates that the relative length of dactyls and propodi is not a phylogenetically significant morphological character for Clibanarius.
To date, there are limited studies on the phylogeny within Clibanarius. Previous phylogenetic analyses of the Anomura and the other genera of diogenid inferred from male reproductive system or molecular markers showed low phylogenetic resolution for interspecific relationships of Clibanarius, in which Clibanarius species were regarded as a supplement (Tudge, 1997; Mantelatto et al., 2006; Tirelli et al., 2008, 2010; Tsang et al., 2011). There were only three researches exclusively explored the molecular phylogeny and morphological characteristics of Clibanarius, all which produced insufficient information on Clibanarius phylogeny and might be caused by limited taxon sampling and inadequate molecular data (Hirose et al., 2010; Negri et al., 2014; Yoshikawa et al., 2018). Our phylogenetic analysis based on all four-gene concatenated dataset presents relatively high support values for more than half of the branch nodes and the major pattern of phylogenetic relationships amongst all studied taxa is consistent across the analytic methods. The present research successfully sheds more lights on the interspecific relationships of this genus. The following interspecific relationships are drawn from analyses of all four-gene concatenated dataset: (ⅰ) C. rutilus and C. infraspinatus show a close relationship, whereas C. snelliusi is suggested to have a sister relationship with them; (ⅱ) C. longitarsus and C. striolatus are shown to be closely related and in accordance with previous studies (Hirose et al., 2010; Yoshikawa et al., 2018). Hirose et al. (2010) and Sha et al. (2015) also predicted this close affinity based on the morphological characters with the dactyl longer than the propodus as well as their habitats; (ⅲ) C. virescens clusters with the group including C. englaucus, C. merguiensis and C. humilis with high support values. The close association of the latter three species is consistent with previous molecular analyses and corroborated by their similarity in general morphology (Osawa and Chan, 2009; Hirose et al., 2010). For example, C. englaucus and C. merguiensis have common characters in dactyl, rostrum, cornea, and ocular acicle, and harbor similar inhabit environments (e.g., rocky shore, coral reef or sea grass bed Sha et al., 2015). The close affinity between C. englaucus and C. humilis was also shown in the tree topologies of Yoshikawa et al. (2018). Furthermore, we find that C. rutilus, C. infraspinatus and C. snelliusi have the telson with barely detectable median cleft, C. longitarsus and C. striolatus have the telson with very slender median cleft, while C. virescens, C. englaucus, C. merguiensis, and C. humilis have the telson with shallow median cleft (McLaughlin et al., 2007; Sha et al., 2015). Thus, we speculate that the morphological characteristic of the median cleft of the telson may be phylogenetically important for Clibanarius. In addition, the remaining two species C. eurysternus and C. corallinus have prominent U-shaped median cleft of the telson, which differs from the other Clibanarius species with shallow or/and slender median cleft. However, no agreement in the branching position of C. eurysternus and C. corallinus is obtained in the current research. The inclusion of more morphological data and extensive taxon collection are necessary to address this question in the future.
5 CONCLUSIONIn this study, we investigate the phylogenetic relationships of Clibanarius based on mitochondrial and nuclear sequence data using a multigene phylogenetic approach. In contrast to Asakura's morphological classification scheme for Clibanarius, the present molecular data provides evidence that the relative length of dactyls and propodi is not phylogenetically significant for Clibanarius. Our study is the first multigene analysis on the phylogeny of Clibanarius and the phylogenetic relationships among some taxa are well resolved. We suggest that (ⅰ) C. snelliusi has a sister relationship with C. rutilus and C. infraspinatus, and the latter two taxa show closer relationships; (ⅱ) C. longitarsus and C. striolatus are shown to be closely related; (ⅲ) C. virescens is divergent from C. merguiensis, C. englaucus, and C. humilis, in which C. merguiensis and C. englaucus are sister groups with C. humilis in our research, reflecting a close relationship among them. We also speculate that the characteristic of the median cleft of the telson may be phylogenetically important for Clibanarius. In addition, our results demonstrate the importance of a large number of nucleotide sites as well as the inclusion of multiple genes with distinct evolutionary histories in the phylogenetic reconstruction of relationships among Clibanarius species. Further studies including more data from genome, extensive sampling and comparison with morphological information are necessary to elucidate phylogenetic relationships within Clibanarius.
6 DATA AVAILABILTY STATEMENTThe datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request. Sequence data that support the findings of this study have been deposited in GenBank under the accession numbers MH508045–MH508087 and MK076123–MK076149.
Ahyong S T, Schnabel K E, Maas E W. 2009. Anomuran phylogeny: new insights from molecular data. In: Martin J W, Crandall K A, Felder D L eds. Decapod Crustacean Phylogenetics. CRC Press, Boca Raton, FL, USA. p.399-414.
|
Asakura A. 2005. Preliminary report of a review of the genus Clibanarius dana, 1852 (Decapoda:Anomura:Diogenidae) from Japan and the adjacent waters. Umiushitsushin, 47: 5-8.
|
Ball E E Jr, Haig J. 1972. Hermit crabs from eastern New Guinea. Pacific Science, 26(1): 87-107.
|
Bracken-Grissom H D, Cannon M E, Cabezas P, Feldmann R M, Schweitzer C E, Ahyong S T, Felder D L, Lemaitre R, Crandall K A. 2013. A comprehensive and integrative reconstruction of evolutionary history for Anomura(Crustacea:Decapoda). BMC Evolutionary Biology, 13: 128.
DOI:10.1186/1471-2148-13-128 |
Brandley M C, Schmitz A, Reeder T W. 2005. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Systematic Biology, 54(3): 373-390.
|
Brandt J F. 1851. Krebse. In: von Middendorf A T ed. Reise in den Äussersten Norden und Osten Sibiriens Während der Jahre 1843 und 1844 Mit Allerhöchster Genehmigung auf Veranstaltung der Kaiserlichen Akademie der Wissenschaften Zu St. Petersburg Ausgeführt und in Verbinding mit Vielen Gelehrten Herausgegeben. Vol. 2, Part 1. St. Petersburg. p.145-147.
|
Brown J M, Lemmon L R. 2007. The importance of data partitioning and the utility of Bayes factors in Bayesian phylogenetics. Systematic Biology, 56(4): 643-655.
DOI:10.1080/10635150701546249 |
Buitendijk A M. 1937. Biological results of the snellius expedition. IV. the paguridea of the snellius expedition. Temminckia, 2: 251-280.
|
Castoe T A, Doan T M, Parkinson C L. 2004. Data partitions and complex models in Bayesian analysis:the phylogeny of Gymnophthalmid. Systematic Biology, 53(3): 448-469.
|
Chen C L, Goy J W, Bracken-Grissom H D, Felder D L, Tsang L M, Chan T Y. 2016. Phylogeny of Stenopodidea(Crustacea:Decapoda) shrimps inferred from nuclear and mitochondrial genes reveals non-monophyly of the families Spongicolidae and Stenopididae and most of their composite genera. Invertebrate Systematics, 30(5): 479-490.
DOI:10.1071/IS16024 |
Cheng J, Gao T X, Miao Z Q, Yanagimoto T. 2011. Molecular phylogeny and evolution of Scomber (Teleostei:Scombridae) based on mitochondrial and nuclear DNA sequences. Chinese Journal of Oceanology and Limnology, 29(2): 297-310.
DOI:10.1007/s00343-011-0033-7 |
Chu K H, Tsang L M, Ma K Y, Chan T Y, Ng P K L. 2009.Decapod phylogeny: what can protein-coding genes tell us? In: Martin J W, Crandall K A, Felder D L eds. Decapod Crustacean Phylogenetics. CRC Press, Boca Raton. p.89-99.
|
Crandall K A, Fitzpatrick J F Jr. 1996. Crayfish molecular systematics:using a combination of procedures to estimate phylogeny. Systematic Biology, 45(1): 1-26.
|
Dana J D. 1851. Conspectus crustaceorum quae in orbis terrarium circumnavigatione, Carolo Wilkes e classe reipublicae foed eratae duce, lexit et descripsit. Proceedings of the Academy of Natural Sciences of Philadelphia, 5: 247-254.
|
Dana J D. 1852. Conspectus crustaceorum, etc., conspectus of the crustacea of the exploring expedition under Capt.Wilkes, U.S.N., including the paguridea, continued, the megalopidea, and the macroura. paguridea, continued, and subtribe megalopidea. Proceedings of the Academy of Natural Sciences of Philadelphia, 6: 6-28.
|
Danforth B N, Brady S G, Sipes S D, Pearson A. 2004. Singlecopy nuclear genes recover cretaceous-age divergences in bees. Systematic Biology, 53(2): 309-326.
|
Danforth B N, Fang J, Sipes S, Brady S G, Almeida E A B, Litman J R, Cardinal S. 2011. Phylogeny and Molecular Systematics of Bees (Hymenoptera:Apoidea). Cornell University, Ithaca, NY.
|
Darriba D, Taboada G L, Doallo R, Posada D. 2012. jModelTest 2:more models, new heuristics and parallel computing. Nature Methods, 9(8): 772.
|
de Grave S, Pentcheff N D, Ahyong S T, Chan T Y, Crandall K A, Dworschak P C, Felder D L, Feldmann R M, Fransen C H J M, Goulding L Y D, Lemaitre R, Low M E Y, Martin J W, Ng P K L, Schweitzer C E, Tan S H, Tshudy D, Wetzer R. 2009. A classification of living and fossil genera of decapod crustaceans. The Raffles Bulletin of Zoology, 21: 1-109.
|
de Haan W. 1833-1850. Crustacea. In: von Siebold P F ed.Fauna Japonica sive Descriptio Animalium, Quae in Itinere per Japoniam, Jussu et Auspiciis Superiorum, qui Summum in India Batava Imperium Tenent, Suspecto, Annis 1823-1830 Collegit, Notis, Observationibus et Adumbrationibus Illustravit. Lugduni-Batavorum, Leiden. p.1-243.
|
de Man J G. 1888. Report on the Podophthalmous Crustacea of the Mergui Archipelago, collected for the Trustees of the Indian Museum, Calcutta, by Dr. John Anderson, F.R.S., Superintendent of the Museum. -Part Ⅳ. Journal of the Linnean Society of London, 22(139): 177-240.
DOI:10.1111/j.1096-3642.1888.tb00031.x |
Excoffier L, Lischer H E L. 2010. Arlequin suite ver 3.5:a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10(3): 564-567.
DOI:10.1111/j.1755-0998.2010.02847.x |
Fabricius J C. 1787. Mantissa insectorum, sistens eorum species nuper detectas adiectis characteribus genericis, differentiis specificis, emendationibus observationibus.Vol. 1. Hafniae: C.G. Proft. xx, p.1-348.
|
Friedlander T P, Regier J C, Mitter C, Wagner D L. 1996. A nuclear gene for higher level phylogenetics:phosphoenolpyruvate carboxykinase tracks mesozoic-age divergences within Lepidoptera (Insecta). Molecular Biology and Evolution, 13(4): 594-604.
DOI:10.1093/oxfordjournals.molbev.a025619 |
Guindon S, Dufayard J F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies:assessing the performance of PhyML 3.0. Systematic Biology, 59(3): 307-321.
|
Hilgendorf F. 1869. Crustaceen. In: van der Decken C C ed.Reisen in Ost-afrika in den Jahren 1859-1865. Leipzig, Heidelberg. p.69-116.
|
Hilgendorf F. 1879. Die von Herrn W. peters in moç ambique gesammelten crustaceen. Monatsberichte der kö niglichen Preussischen Akademie des Wissenschaften zu Berlin, 1878: 782-851.
|
Hirose M, Osawa M, Hirose E. 2010. DNA barcoding of hermit crabs of genus Clibanarius Dana, 1852 (Anomura:Diogenidae) in the Ryukyu Islands, southwestern Japan. Zootaxa, 2414(1): 59-66.
DOI:10.11646/zootaxa.2414.1.5 |
Huelsenbeck J P, Ronquist F. 2001. MrBayes:Bayesian inference of phylogeny. Bioinformatics, 17: 754-755.
DOI:10.1093/bioinformatics/17.8.754 |
Knowlton N, Mills D K. 1992. The systematic importance of color and color pattern:evidence for complexes of sibling species of snapping shrimp (Caridea:Alpheidae:Alpheus)from the Caribbean and Pacific coasts of Panama. Proceedings of the San Diego Society of Natural History, 18: 1-5.
|
Krauss F. 1843. Die Sudafrikanischen Crustaceen. Eine Zusammenstellung aller bekannten Malacostraca.Bemerkungen über Deren Lebensweise und Geographische Verbreitung, Nebst Beschreibung und Abbildung Mehrerer Neuen Arten. E. Schweizerbart'sche Verlagsbuchhandlung, Stuttgart. p.68.
|
Leys R, Cooper S J B, Schwarz M P. 2002. Molecular phylogeny and historical biogeography of the large carpenter bees, genus Xylocopa (Hymenoptera:Apidae). Biological Journal of the Linnean Society, 77(2): 249-266.
DOI:10.1046/j.1095-8312.2002.00108.x |
Li C P, de Grave S, Chan T Y, Lei H C, Chu K H. 2011. Molecular systematics of caridean shrimps based on five nuclear genes:implications for superfamily classification. Zoologischer Anzeiger-A Journal of Comparative Zoology, 250(4): 270-279.
DOI:10.1016/j.jcz.2011.04.003 |
Malay M C D, Rahayu D L, Chan T Y. 2018. Hermit crabs of the genera Calcinus Dana, Clibanarius Dana, and Dardanus Paul'son from the PANGLAO 2004 Expedition, with description of a new species and a checklist of the hermit crabs of the Philippines (Crustacea:Anomura:Paguroidea). The Raffles Bulletin of Zoology, 66: 23-65.
|
Mantelatto F L M, Robles R, Biagi R, Felder D L. 2006. Molecular analysis of the taxonomic and distributional status for the hermit crab genera Loxopagurus Forest, 1964 and Isocheles Stimpson, 1858 (Decapoda, Anomura, Diogenidae). Zoosystema, 28(2): 495-506.
|
McLaughlin P A, Komai T, Lemaitre R, Rahayu D L. 2010. Annotated checklist of anomuran decapod crustaceans of the world (exclusive of the Kiwaoidea and families Chirostylidae and Galatheidae of the Galatheoidea) Part Ⅰ-Lithodoidea, Lomisoidea and Paguroidea. The Raffles Bulletin of Zoology, 23: 5-107.
|
McLaughlin P A, Murray T. 1990. Clibanarius fonticola, New species (Anomura:Paguridea:Diogenidae), from a freshwater pool on Espiritu Santo, Vanuatu. Journal of Crustacean Biology, 10(4): 695-702.
DOI:10.2307/1548413 |
McLaughlin P A, Rahayu D L, Komai T, Chan T Y. 2007. A Catalog of the Hermit Crabs (Paguroidea) of Taiwan. National Taiwan Ocean University, Keelung, China.
|
Meyer C P. 2003. Molecular systematics of cowries(Gastropoda:Cypraeidae) and diversification patterns in the tropics. Biological Journal of the Linnean Society, 79(3): 401-459.
DOI:10.1046/j.1095-8312.2003.00197.x |
Milne Edwards P M. 1848. Note sur quelques nouvelles espèces du genre Pagure. In: Annales des Sciences Naturelles-Zoologie et Biologie Animale. Paris. p.59-64.
|
Minegishi Y, Aoyama J, Inoue J G, Miya M, Nishida M, Tsukamoto K. 2005. Molecular phylogeny and evolution of the freshwater eels genus Anguilla based on the whole mitochondrial genome sequences. Molecular Phylogenetics and Evolution, 34(1): 134-146.
DOI:10.1016/j.ympev.2004.09.003 |
Moritz C, Dowling T E, Brown W M. 1987. Evolution of animal mitochondrial DNA:relevance for population biology and systematics. Annual Review of Ecology and Systematics, 18: 269-292.
DOI:10.1146/annurev.es.18.110187.001413 |
Moriyama E N, Powell J R. 1997. Synonymous substitution rates in Drosophila:mitochondrial versus nuclear genes. Journal of Molecular Evolution, 45(4): 378-391.
|
Negri M, Lemaitre R, Mantelatto F L. 2014. Molecular and morphological resurrection of Clibanarius symmetricus(Randall, 1840), a cryptic species hiding under the name for the "Thinstripe" hermit crab C. vittatus (Bosc, 1802)(Decapoda:Anomura:Diogenidae). Journal of Crustacean Biology, 34(6): 848-861.
DOI:10.1163/1937240X-00002277 |
Ortmann A E. 1892. Die decapoden-krebse des strassburger museums, mit besonderer berücksichtigung der von herrn dr. döederlein bei Japan und bei den Liu-Kiu-Inseln gesammelten und zur zeit im strassburger museum auf bewahrten formen. Ⅳ Theil. Die abtheilungen galatheidea und paguridea. Zoologische Jahrbücher, 6: 241-325.
DOI:10.5962/bhl.part.26455 |
Osawa M, Chan T Y. 2009. Additional records of hermit crabs(Crustacea:Decapoda:Anomura:Paguroidea) from Taiwan. Proceedings of the Biological Society of Washington, 122(3): 317-332.
DOI:10.2988/08-50.1 |
Palero F, Crandall K A. 2009. Phylogenetic inference using molecular data. In: Martin J W, Crandall K A, Felder D L eds. Decapod Crustacean Phylogenetics. CRC Press, Boca Raton. p.67-88.
|
Rahayu D L. 1999. Descriptions of two new species of hermit crabs, Clibanarius rubroviria and C. rutilus (Crustacea:Decapoda:Anomura:Diogenidae) from Indonesia. The Raffles Bulletin of Zoology, 47(2): 299-307.
|
Roterman C N, Lee W K, Liu X M, Lin R C, Li X Z, Won Y J. 2018. A new yeti crab phylogeny:vent origins with indications of regional extinction in the East Pacific. PLoS One, 13(3): e0194696.
DOI:10.1371/journal.pone.0194696 |
Sha Z L, Xiao L C, Wang Y L. 2015. Study on the Taxonomy of the Family Diogenidae (Crustacea:Decapoda:Anomura:Paguroidea) from China Seas. Science Press, Beijing, China. p.1-252.
(in Chinese)
|
Swofford D L. 2002. PAUP*:Phylogenetic Analysis Using Parsimony (and Other Methods). Sinauer Associates, Sunderland, MA.
|
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6:molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2725-2729.
DOI:10.1093/molbev/mst197 |
Tirelli T, Pessani D, Silvestro D, Tudge C. 2008. Reproductive biology of Mediterranean hermit crabs:fine structure of spermatophores and spermatozoa of Diogenes pugilator(Decapoda:Anomura) and its bearing on a sperm phylogeny of Diogenidae. Journal of Crustacean Biology, 28(3): 534-542.
DOI:10.1651/07-2956.1 |
Tirelli T, Silvestro D, Pessani D, Tudge C C. 2010. Description of the male reproductive system of Paguristes eremita(Anomura, Diogenidae) and its placement in a phylogeny of diogenid species based on spermatozoal and spermatophore ultrastructure. Zoologischer Anzeiger-A Journal of Comparative Zoology, 248(4): 299-312.
DOI:10.1016/j.jcz.2010.01.001 |
Toon A, Finley M, Staples J, Crandall K A. 2009. Decapod phylogenetics and molecular evolution. In: Martin J W, Crandall K A, Felder D L eds. Decapod Crustacean Phylogenetics. CRC Press, Boca Raton. p.15-29.
|
Tsang L M, Chan T Y, Ahyong S T, Chu K H. 2011. Hermit to King, or Hermit to all:multiple transitions to Crab-like forms from Hermit crab ancestors. Systematic Biology, 60(5): 616-629.
DOI:10.1093/sysbio/syr063 |
Tsang L M, Ma K Y, Ahyong S T, Chan T Y, Chu K H. 2008. Phylogeny of Decapoda using two nuclear protein-coding genes:origin and evolution of the Reptantia. Molecular Phylogenetics and Evolution, 48(1): 359-368.
DOI:10.1016/j.ympev.2008.04.009 |
Tsang L M, Schubart C D, Ahyong S T, Lai J C Y, Au E Y C, Chan T Y, Ng P K L, Chu K H. 2014. evolutionary history of true crabs (Crustacea:Decapoda:Brachyura) and the origin of freshwater crabs. Molecular Biology and Evolution, 31(5): 1173-1187.
DOI:10.1093/molbev/msu068 |
Tudge C C. 1997. Phylogeny of the Anomura (Decapoda, Crustacea):spermatozoa and spermatophore morphological evidence. Contributions to Zoology, 67(2): 125-141.
DOI:10.1163/18759866-06702002 |
Wiegmann B M, Mitter C, Regier J C, Friedlander T P, Wagner D M, Nielsen E S. 2000. Nuclear genes resolve Mesozoicaged divergences in the insect order Lepidoptera. Molecular Phylogenetics and Evolution, 15(2): 242-259.
DOI:10.1006/mpev.1999.0746 |
Williams J D, McDermott J J. 2004. Hermit crab biocoenoses:a worldwide review of the diversity and natural history of hermit crab associates. Journal of Experimental Marine Biology and Ecology, 305(1): 1-128.
DOI:10.1016/j.jembe.2004.02.020 |
Yoshikawa A, Nakano T, Satoh T P, Asakura A. 2018. A colour variation of Clibanarius virescens (Krauss, 1843)(Decapoda, Anomura) collected from Amami Oshima Island and Okinawa, Japan. Crustaceana, 91(1): 85-101.
DOI:10.1163/15685403-00003748 |
 2019, Vol. 37
2019, Vol. 37