Institute of Oceanology, Chinese Academy of Sciences
Article Information
- JIANG Yan, LIU Xuezhou, XU Yongjiang, SHI Bao, WANG Bin
- Microbiota characteristics in Sebastes schlegelii intestine in early life stages
- Journal of Oceanology and Limnology, 38(1): 275-287
- http://dx.doi.org/10.1007/s00343-019-9011-2
Article History
- Received Jan. 29, 2019
- accepted in principle Apr. 3, 2019
- accepted for publication May. 3, 2019
2 Laboratory for Marine Fisheries and Food Production Processes, Pilot National Laboratory for Marine Science and Technology(Qingdao), Qingdao 266237, China
The intestinal tract of animal is a very complex ecological system, in which a diversiform and dynamical microbial community was included. The composition and activity of this microflora were closely related to the genetic information, lifestyle, nutrient supply, and external environment of the host (Li et al., 2012; Nicholson et al., 2012). Microbes adhere to the specific sites in the gut of host and live on it. However, they could accomplish many physiological functions for the host (Forsythe and Bienenstock, 2010; Pérez et al., 2010; Karasov et al., 2011), such as nutrition and immunity, to prevent pathogenic bacteria from settlement, to balance the energy and keep the normal mucosal immunity, and so on (Gallo and Nakatsuji, 2011; Kim et al., 2013; Ye et al., 2014). Moreover, the co-action between gut microbiota and animal endocrine system can affect the host behavior and physiological activity (Gacias et al., 2016; Ntranos and Casaccia, 2018).
In recent years, with the development of microbiology and molecular ecology, the composition and structural characteristics of the intestinal microbial community for animals attract much more attention of researchers in the world. More and more studies are reported on gut microflora (Ghanbari et al., 2015; Dawood and Koshio, 2016; Zhao et al., 2016; Jiang et al., 2019). Many types of factors affect the successful colonization of microbes in the fish gut. Reports have pointed out that the fish could influence the intestinal microflora composition of themselves. In addition, nutritional conditions, different physiological phases, feeding habits of host, and environmental factors can affect the adhesion of microbes in intestine (Horne and Baxendale, 1983; Ringø et al., 2003; Tanaka et al., 2009; Kelly, 2010; Sanchez et al., 2012; Sullam et al., 2012; Banerjee and Ray, 2017). Consequently, it is impossible that external microflora can successfully colonize on the intestinal tract of the host and play the physiological functions (Chung et al., 2012; Ferreira and Veldhoen, 2012).
Fish is the most diverse group of vertebrates on earth, and they live in water, which caused much more environmental factors could affect their gut microbial community (Tanaka et al., 2009; Kelly, 2010; Sullam et al., 2012; Banerjee and Ray, 2017; Jiang et al., 2019). Therefore, the fish gut microbiota is more dynamic compared with the terrestrial animal. In China, black rockfish (Sebastes schlegelii) is one of the key commercial aquaculture fish. There are several factors that restrict larvae growth, such as cannibalism, bacteriosis, and so on (Kitani et al., 2008; Liu et al., 2016). The stable supply of larvae with high quality is the primary assurance for the healthy and sustained development of this industry. At present, the study on the physiological health of black rockfish larvae is relatively few, whereas the systematic research of larval intestinal physiology during the developmental stage is rare. Under these circumstances, the research and application of indigenous probiotics for black rockfish will be severely restricted.
In this study, we researched the composition and main influential factors of microflora, the abundance variation trend and resource of some core microbiota in black rockfish larval and juvenile intestine during the developmental stage. Based on this, the characteristics of the microflora structure in the gut were ascertained, and the process of intestinal microflora succession in the phase of breeding was revealed. Meantime, we analyzed the relativity of microbial community between larvae and external environment including water and feed. Our findings may provide basic data for exploring indigenous probiotics, which can improve the digestive and absorptive capacity and the immunity of black rockfish.
2 MATERIAL AND METHOD 2.1 The breeding management for larvae and juvenilesThe breeding experiment was performed in a black rockfish hatchery located in Qingdao, Shandong Province, China. All larvae were hatched from the same batch fertilized eggs and distributed in nursery ponds (5.0 m×5.0 m×1.0 m). Experimental fish from three random ponds were classified as parallel groups. The breeding management strategy was as follow: [NH4+-N] was lower than 0.1 mg/L, the temperature was 18–20℃, salinity was 30, dissolved oxygen was higher than 5 mg/L.
On day 3 after hatching (DAH), black rockfish larvae started the first feeding food-rotifer (Branchionus plicatilis). On 3–15 DAH, larvae were fed on rotifer, 12–45 DAH were brine shrimp (Artemia sinica); and formulated feed was gradually added after 40 days. The breeding process was strictly according to the practices employed in the black rockfish larval breeding hatchery.
2.2 Samples collection and processAll intestine, water, and feed samples were collected on 1 DAH, 9 DAH, 20 DAH, 54 DAH, and 95 DAH, except for the feed sample on 1 DAH. The process of samples referred to Jiang et al. (2019).
The sampling regime for intestine was: on 1 DAH and 9 DAH, 50 morphologically normal larvae from each pond were randomly collected, 30 larvae on 20 DAH, 10 juveniles on 54 DAH and 5 juveniles on 95 DAH. All selected fish were starved for 12 h and washed three times with sterile seawater for 20 min each time. After the narcotization using MS-222 (Fluka, USA) and wiping with 75% alcohol, the larvae and juveniles were dissected strictly according to the regulations from local government and the Institutional Animal Care & Use Committee (IACUC) of Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences. Under the sterile condition, the intestine was taken out, the content in the intestine was removed and the intestine was washed with sterile saline. Then, the intestines were stored in liquid nitrogen, respectively. The entire larva on 1 DAH was retained because the differentiation of gut was not obvious. All intestinal samples were marked with G1, G9, G20, G54, and G95 for larvae or juveniles guts on 1 DAH, 9 DAH, 20 DAH, 54 DAH, and 95 DAH, respectively. While G1.1, G1.2, and G1.3 represented three replicates on 1 DAH, and the same marking method was used for the other samples.
In addition, three wild black rockfish adults (body weight was 205±32 g) were collected from Yellow sea around the hatchery location where the breeding experiment was conducted and marked with G-W1, G-W2, and G-W3, respectively. The process of wild adult samples is the same to those for the larvae and juveniles samples.
Water samples: at each sampling point, 10 L water was collected from each pond and filtrated with a 0.22-μm membrane. Membranes were stored in liquid nitrogen and used to extract total DNA of microbes. Water samples were labeled with W1, W9, W20, W54, and W95, respectively.
Feed samples: rotifer and brine shrimp were sampled on 9 DAH and 20 DAH, respectively. Rotifer or brine shrimp was used to feed larvae and a juvenile after nutrient enrichment. The formulated feed (Haitong, Weifang Santong Group, China) sampled on 54 DAH and 95 DAH. The live feed was filtered and washed with the sterile condition; about 5 grams of rotifer and brine shrimp were sampled and then stored in liquid nitrogen for DNA extraction. The samples were labeled with F9, F20, F54, and F95, which represented the feed used on 9 DAH, 20 DAH, 54 DAH, and 95 DAH, respectively.
2.3 Total DNA extraction and sequencingThe larval and juvenile samples of each replicate at each sampling point were each stirred and homogenized. Three equal-weight homogenates from three breeding ponds as parallel groups and extracted total DNA for microbes using the QIAamp DNA mini kit (QIAGEN, Germany) according to the manufacturer's instructions. Membranes were cut into pieces and extracted total DNA for microbes in water samples using E.Z.N.A.® Soil DNA Kit (Soil DNA Kit D5625, Omega Biotek, USA). All feed samples in equal weight were extracted total DNA for microbes with the QIAamp DNA mini kit.
The V3 and V4 regions of 16S rDNA were amplification through a polymerase chain reaction, and the primer was: 343F (5′-TACGGRAGGCAGCAG-3′) and 798R (3′- AGGGTATCTAATCCT-5′) (Nossa et al., 2010). After the amplified, DNA was confirmed with agarose gel electrophoresis, and high throughput sequencing was conducted with an Illumina MiSeq PE300 system.
2.4 Data processing and analysisRaw data were split with Trimmomatic (v 0.35) (Bolger et al., 2014), spliced with Flash (v 1.2.11) (Reyon et al., 2012), removed the chimera with Uchime (v. 4.2) (Edgar et al., 2011), and so forth after obtaining from high throughput sequencing. We get the total effective tags of all samples, which were clustered using Vsearch (v 2.4.2) (Rognes et al., 2016). Several effective tags were clustered into one operational taxonomic unit (OTU) while their shared sequencing identity was higher than 97%. Representative sequences of OTUs were selected and annotated in species using RDP Classifier (v. 2.2) (Wang et al., 2007) and Silva database (v. 123) (Quast et al., 2013). Data for all samples were homogenized basing on the standard for the sample with the least data. Alpha and beta diversity analyses were performed based on this homogenization process.
The analysis of variance (ANOVA) of microflora structure for samples was carried out using SPSS 17.0 software (IBM, USA). The significant level was set at P < 0.05 and the data was expressed as means±standard error.
3 RESULTThe barcode and primer sequences were removed, and the unqualified tags and chimera sequences were cleaned for the raw data obtained from 42 samples using the Illumina MiSeq PE300 system. Thus, the effective tags were obtained for use in the subsequent analysis. Each sample had an average of 25 766 effective tags.
The sequencing results showed that the G1 larvae possess the most OTU numbers and bacteria groups at phylum, class and other levels, while the G54 juveniles have the most bacteria groups at family and genus levels (Table S1).
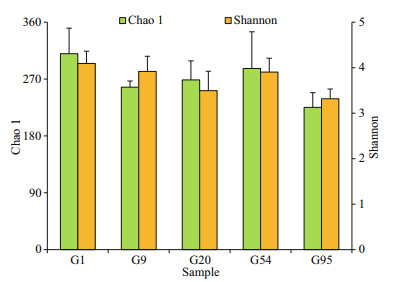
3.1 Diversity and taxonomic composition of microflora in the intestineThroughout the whole early life stage of black rockfish, the Chao 1 value initially decreased and then increased, but there was a sharp decline after the G54 stage (Fig. 1). The highest Chao 1 value (310.3) was obtained on 1 DAH, whereas no significant differences were detected in all samples (P > 0.05). The highest Shannon index was also observed in G1 larvae (4.1), and showed a descending trend (P > 0.05) through the experimental period (Fig. 1).
|
| Fig.1 Chao 1 and Shannon index of microbiota in larval and juvenile intestine of black rockfish Values with different letters differed significantly on the same day (n=3, P < 0.05). G1 represents the larvae on 1 DAH, G9 and G20 represent larval intestinal samples on 9 DAH and 20 DAH, while G54 and G95 represent juvenile guts on 54 DAH and 95 DAH, respectively. |
The top-ten phyla in each intestinal sample are shown in Fig.S1. The dominant phylum was Proteobacteria, and the relative abundance for each sample was 72.17%–92.08% (mean=80.64%), following by the Firmicutes with the abundance was 5.96%–20.73%. Likewise, the wild fish intestine was also numerically dominated by Proteobacteria. However, Firmicutes, Cyanobacteria, and Tenericutes were also the important phyla groups in the wild fish intestine.
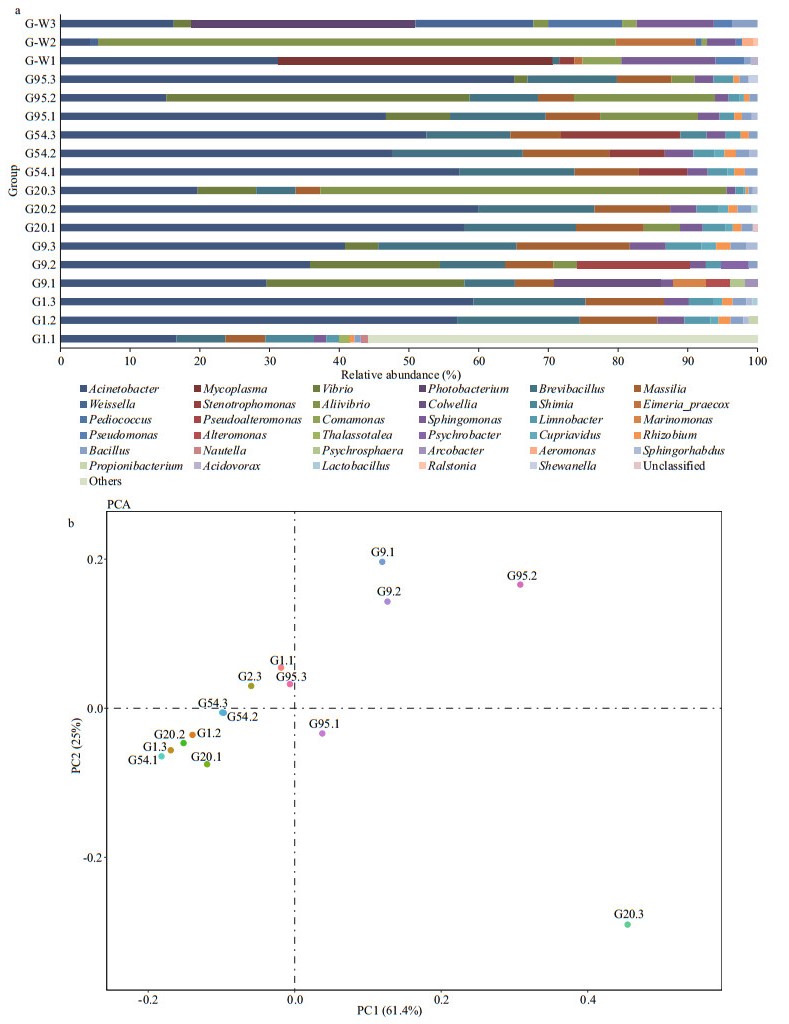
The ten most abundant OTUs within the different samples were determined to understand further the important bacteria at the genus level (Fig. 2a). Acinetobacter was the most abundant, accounting for 36.10% of the total. The lowest abundance was 26.19% while the larvae were fed on rotifer, and the abundance of Acinetobacter reached the highest value (45.28%) when the juveniles were fed with formulated feed on 54 DAH. Brevibacillus, Massilia, and Sphingomonas were also the dominant genera in black rockfish larval and juvenile guts. The abundance of these three genera was 13.76%, 9.81%, and 3.17% before the first feeding, respectively. However, their abundances all declined from 9 to 95 DAH. Vibrio became the dominant species after the first feeding, and the highest abundance (12.41%) appeared on 9 DAH. Compared with the artificial breeding larvae and juveniles, Acinetobacter, Mycoplasma, Photobacterium, Weissella, Aliivibrio, and Sphingomonas were the dominant genera in wild fish guts. The samples distributed around the larvae on 1 DAH except for some replicates on 9, 20, and 95 DAH (Fig. 2b). Likewise, distributions of microflora in the gut were gradually changed in different feed stage.
|
| Fig.2 The structural characteristic of microflora in black rockfish larval and juvenile samples a. relative abundances of the main genus; b. principal components analysis (PCA) based on the operational taxonomic unit (OTU) level. G-W1, G-W2, and G-W3 are the three intestinal samples for wild black rockfish. G1 represents the larvae on 1 DAH, G9 and G20 represent larval intestinal samples on 9 DAH and 20 DAH, while G54 and G95 represent juvenile guts on 54 DAH and 95 DAH, respectively. G1.1, G1.2, and G1.3 represented three replicates on 1 DAH, and the same as others. |
The shared species at the genus level in fish gut samples are presented in Fig. 3. After the first feeding, the numbers of shared genera among larval and juvenile intestines on 9 to 95 DAH was 34 while 32 shared genera among the total species in all larval samples. Analyzing the top-ten genera of each sample and this shared microflora, we got the main microflora for healthy larval guts including Propionibacterium, Lactobacillus, Bacillus, Vibrio, Acinetobacter, Brevibacillus, Sphingomonas, Massilia, Stenotrophomonas, Rhizobium, Sphingorhabdus, Cupriavidus, Pseudoalteromonas, and Limnobacter. Abundances of genera in Fig. 4a & b presented the trend of increased initially and then decreased along with the growth of larvae except that on 1 DAH, while trends of Vibrio and another main genus was similar to "V" and reversed "N" in Fig. 4c & d, respectively. However, at the whole scale, abundances of Propionibacterium, Lactobacillus, Acinetobacter, and Vibrio in larval guts on 9 DAH were lower than those on 95 DAH, and the others showed the opposite order according to the abundances.
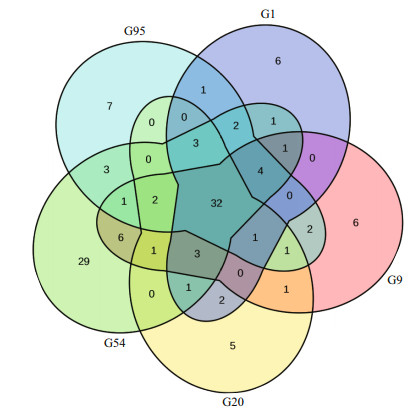
|
| Fig.3 The shared genus among different stages for black rockfish larval and juvenile gut samples The species numbers in each group was the number of shared species among three replicates in this testing. |

|
| Fig.4 The variation trend of main microflora abundance in black rockfish intestines during the developmental stage |
Therefore, the microflora structures in guts gradually changed with the growth of black rockfish larvae. However, the dominant genera, such as Acinetobacter, Vibrio, Brevibacillus, Massilia, Stenotrophomonas, Pseudoalteromonas, Sphingomonas, Bacillus, Lactobacillus, etc., always colonized in the intestines. The composition of major species existed in larval and juvenile intestines was obviously different from that in wild ones.
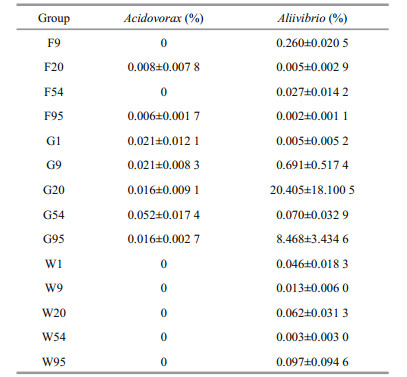
3.2 The relativity between intestinal microbiota and environmental microbiotaThrough the comparison, Acidovorax and Aliivibrio were the shared species in larval intestines on 9 to 95 DAH, but not included 1 DAH (Table S2). We found Acidovorax existed in all intestinal samples except one replicate on day 1, and one feed sample on 20 DAH and all feed samples on 95 DAH. There were no one water samples contained Acidovorax (Table S3).
The shared species among three replicates of different types of samples and inter-group on each collecting day at the genus level are presented in Fig. 5a. Averaged numbers of the shared genus in guts, water, and feed were 63.6, 80.2, and 148.5. The minimum value of shared species in feed samples was higher than the maximum value in intestines or water. Based on these, averaged numbers of shared genus between guts and water, guts and feed were 20 and 33.25, while 9–19 (mean=15) shared genera among these three samples on each sampling day. Additionally, these shared species between guts and water, guts and feed on each sampling day are shown in Table S4. Based on these results, we found that the shared species among all larval samples (32) were higher than those in the feed (22) or water (22) (Fig. 5b). However, there were only four shared species among these three types of samples that are consisted of Bacillus, Lactococcus, Pseudoalteromonas, and Vibrio (Table 1). Compared with these 4 shared genera, the specific and shared species between guts and feed were Lactobacillus, Acinetobacter, and Pseudomonas, while only Glaciecola between guts and water. Moreover, Lactococcus and Glaciecola were the main genera in feed and water while Pseudomonas only dominant in the feed. Abundances of the other five shared species were all top-ten in larval intestines according to relative abundance (Figs. 2a, S2).
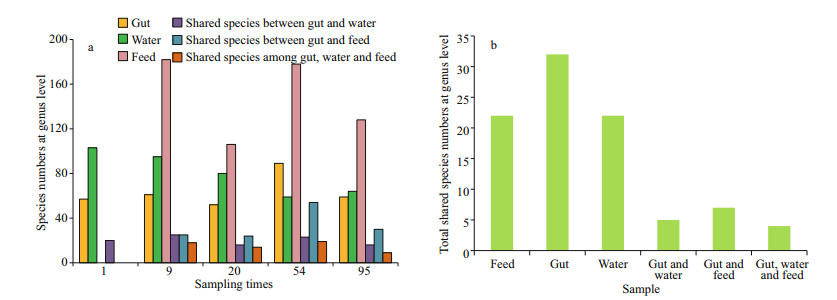
|
| Fig.5 Numbers of shared genera among different samples along the developmental stage for black rockfish a. at each sampling DAH in one, two and three kinds of sample; b. in different types of sample in all five stages. |
The unweighted pair group method with arithmetic mean (UPGMA) based on weighted UniFrac distances showed that the microbial community structure of G9 and G95 are all closed to those of F9 and F95 (Fig. 6). However, the microbiota composition of W20, W54, F20, and F54 could influence that of G20 and G54.
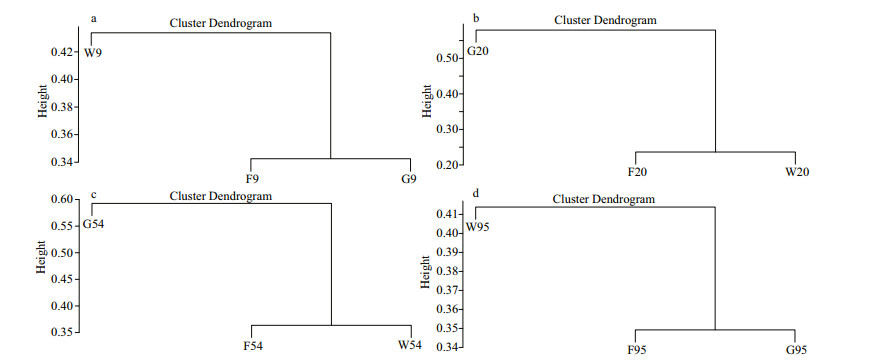
|
| Fig.6 Clustering analysis with unweighted pair group method with arithmetic mean (UPGMA) for the microbiota in the fish gut, breeding water and feed a, b, c and d respectively represent samples on 9, 20, 54 and 95 DAH. |
Therefore, Acidovorax, Aliivibrio, Bacillus, Lactococcus, Pseudoalteromonas, Vibrio, Lactobacillus, Acinetobacter, Pseudomonas and Glaciecola were the core genera of this study during the larval developmental stage. Compared with breeding water, microbiota structure in feed was more similar to that in larval and juvenile guts.
3.3 Core microbiotaRelative abundances of Acidovorax and Aliivibrio in all samples are shown in Table 2. Changes of abundances for Acidovorax in larval intestines were not obvious, and percentages of Acidovorax in feed samples were rather low. The changes in abundances for Aliivibrio in intestines showed the increasing trend as a whole. Aliivibrio became the top-ten genus in guts on 20 DAH (20.405%) and 95 DAH (8.468%) in Fig. 2a. Abundances of Aliivibrio in water and feed samples presented irregular changes and were lower than 0.3%.
Relative abundances of another 8 shared species in inter-group were presented in Fig. 7. In intestinal samples, Bacillus, Acinetobacter and Pseudomonas were dominant genera with higher abundance. Trends of these three species were close along with the growth of larvae and juveniles, and similar with Lactobacillus, Lactococcus, and Glaciecola, but obviously opposite to Vibrio and Pseudoalteromonas (Fig. 7a). However, abundances of Bacillus, Lactobacillus, Lactococcus, and Vibrio increased initially and then decreased, Acinetobacter and Pseudomonas presented the increasing trend in different kinds of feed samples (Fig. 7b). Bacillus, Lactobacillus, Lactococcus, Pseudoalteromonas, Acinetobacter, and Pseudomonas presented the descending trend in water samples during the developmental stage of black rockfish larvae and juveniles (Fig. 7c).
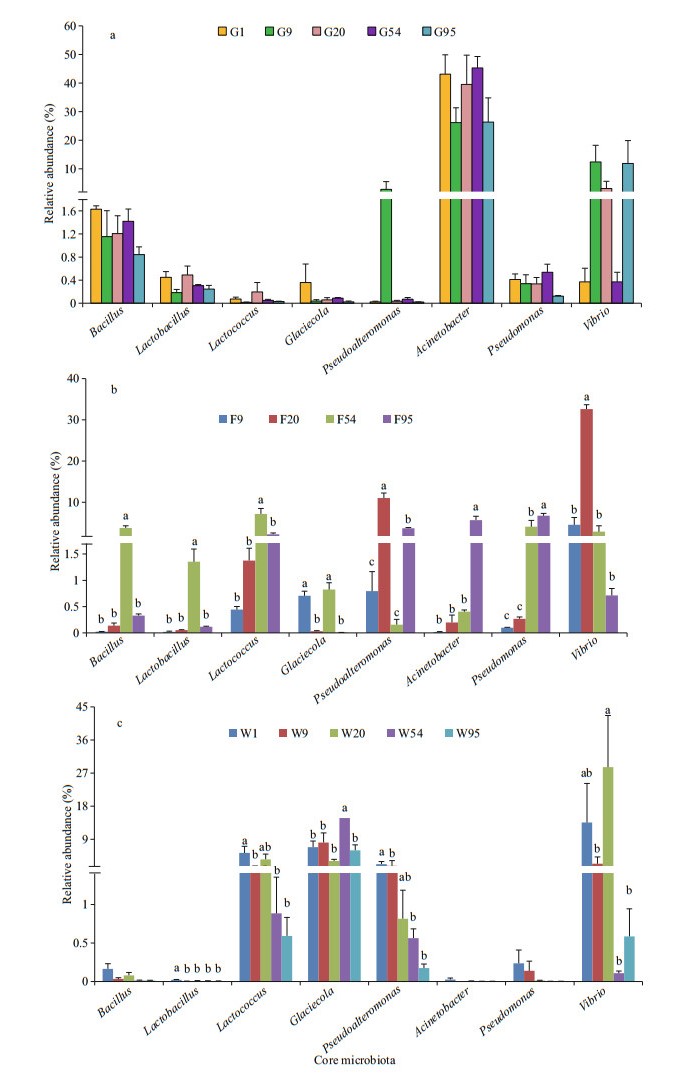
|
| Fig.7 The trends of abundances of shared genera on each sampling day a. in black rockfish intestines; b. in various feed samples; c. in breeding water. |
Proteobacteria with the highest abundance and Firmicutes are the two dominant phyla in normal larval guts during the whole developmental stage in this study. The same results were obtained in rainbow trout and turbot larvae and juveniles, but in the genus level, there are some differences (Desai et al., 2012; Jiang et al., 2019). This disparity is caused by the differences of species, dietary habits, living environment, and the functions of microbiota (Banerjee and Ray, 2017). After the analysis of shared and dominant genera, we get the major microflora always living in larval guts. Abundances of almost major microflora were decreased from new hatched larval stage to rotifer feed stage. In later developmental stage for black rockfish, this microflora presented increased initially and then decreased trend. These might be caused by the increasing uniformity of microbiota with the feeding and growth. However, Vibrio showed the opposite trend, which was coeffected by microbiota in feed and breeding water. Relative studies suggested environmental and ecological factors could influence the gut bacterial communities of fish (Tanaka et al., 2009; Kelly, 2010; Sullam et al., 2012; Shabat et al., 2016; Zhang et al., 2016; Banerjee and Ray, 2017; Jiang et al., 2019).
Analyzing the common genus among all samples, we obtained the core microbiota at the genus level for this study, including Acidovorax, Aliivibrio, Bacillus, Acinetobacter, Pseudomonas, Lactobacillus, Lactococcus, Glaciecola, Vibrio, and Pseudoalteromonas. In these shared species of all samples in each stage, Aliivibrio, Bacillus, Acinetobacter, Lactobacillus, Vibrio, and Pseudoalteromonas were the top-ten microbiota according to the relative abundances in larval intestines. Additionally, we noted that Acidovorax existed in guts on 1 DAH, while the larvae did not possess the feeding behavior and few feed samples, which suggested that genus Acidovorax came from the parents of black rockfish larvae and/or fertilization process and could always colonize in gut, though the abundances were lower than 0.1%. Glaciecola and Pseudomonas were also investigated in larval samples on 1 DAH and the water samples, inferring these two genera from the parents of black rockfish larvae and/or fertilization process. Meanwhile, the dominant genera Lactobacillus and Acinetobacter were the two shared species among all intestinal samples, which suggested that they mainly came from the parents of black rockfish larvae and/or fertilization process and could successfully and persistently colonize in larval gut. Moreover, Lactobacillus and Acinetobacter presented in all feed samples, illustrating that feed was the main effective factor in abundance after the first feeding. It is likely that feed was the major element influencing abundances of Acinetobacter, Pseudomonas, Lactobacillus and Bacillus via the characteristics of changes in relative abundances (Fig. 7). The variation trend of abundances of Glaciecola was similar to that in feed, but with higher abundances in water, suggesting feed and water could co-affect the existence of this shared genus in gut. Additionally, the differences of dominant species in guts between the artificial breeding larvae and the wild ones implied that the living environment could obviously affect the microflora structure in black rockfish guts. These illustrated that feed and water could influence the dynamic balance of intestinal microbiota. The relative results will point out the direction for us in the subsequent research. The previous study indicated that nutritional components and types of feed could affect the microbial community in fish larval guts (Banerjee and Ray, 2017; Li et al., 2017b), but it is impossible that microbiota in live feed could obviously change larval gut microflora structure (Bakke et al., 2013). These different results might relate to the type of fish, physiological characters and their living environment (Tanaka et al., 2009; Kelly, 2010; Sullam et al., 2012; Shabat et al., 2016; Zhang et al., 2016; Banerjee and Ray, 2017; Jiang et al., 2019).
Some strains belonging to Acidovorax are the dangerous bacteria for plants (Adhikari et al., 2017; Yan et al., 2017), but rarely present as the dominant species and no reports point the pathogenicity in aquatic animals (Desai et al., 2012; Chun et al., 2017). Some Glaciecola strains have been reported to produce lipopolysaccharide (LPS) and other bioactive compounds (Baik et al., 2006; Qian and Xu, 2007), which are seen as an effective stimulant for the immune system in various fish (Swain et al., 2008). Chen et al. (2010) indicated that LPS extracted from Glaciecola polaris, a nonpathogenic strain, could improve the innate immunity of Lateolabrax japonicas. Therefore, the genera Glaciecola and Acidovorax colonized in larval and juvenile guts might be treated as nonpathogenic bacteria.
Certainly, many beneficial species always adhered on the larval guts and participated in maintaining the balance of intestinal microbiota through competition for nutrients, product inhibition, and some other interactions. Bacillus, Lactobacillus and Lactococcus are usually regarded as the probiotics in aquaculture (Urdaci et al., 2004; Ziaei-Nejad et al., 2006; Chantharasophon et al., 2011; Pérez-Sánchez et al., 2014). Some Bacillus could effectively regulate the microflora structures of B. plicatilis and A. sinica (Jiang et al., 2018). Richards et al. (2017) isolated three pigmented Pseudoalteromonas strains from seawater, which could inhibit the growth of marine pathogens and even kill them, including Vibrio vulnificus, Vibrio parahaemolyticus, Vibrio cholerae, Photobacterium damselae, and Shewanella algae, through the secretion of proteolytic enzymes having antimicrobial properties. Leyton et al. (2017) pointed Pseudoalteromonas sp. as a probiotic introducing through living feed (rotifers and Artemia sp.) could increase the larval survival rate of Seriola lalandi. However, few strains belonging to Pseudoalteromonas could cause Montipora white syndrome in Montipora capitata (Beurmann et al., 2017).
Aliivibrio and Vibrio are commonly considered as the causative agents of vibriosis in marine aquaculture (Hjerde et al., 2008; Karlsen et al., 2014; Muñoz-Atienza et al., 2014). V. alginolyticus, V. harveyi, and V. parahaemolyticus are the representative pathogens (Alcaide et al., 1999; Zorrilla et al., 2003; Liu et al., 2004; Austin and Zhang, 2006). Several species in genus Pseudomonas are known as the pathogenic bacteria for effect various marine cultured fish, such as turbot (López-Romalde et al., 2003; Magi et al., 2009) and cod (Ferguson et al., 2004). Working as a grave pathogen for human, genus Acinetobacter gathers more eyes from medical researchers. To date, many strains in Acinetobacter sp., such as A. lwoffii, A. baumannii, A. johnsonii, A. pittii, A. schindleri, and A. calcoaceticus, have been obtained from cultured fish around the world, and even become the multi-drug resistant fish pathogens (Reddy and Mastan, 2013; Kozińska et al., 2014; Li et al., 2017a; Lynch III et al., 2017; Wong et al., 2017). These four harmful genera, especially Vibrio and Acinetobacter, with higher abundances always adhere to the normal larval and juvenile gut in this study. Moreover, in larval and juvenile breeding stage, the immune system function of larvae is defective. Once abundances of these bacteria are increased to break the balance of gut microbiota in some condition, which will cause diseases in aquaculture and the infection will be quickly extended for the rapid reproduction of bacteria, and the loss will not be estimated (Verschuere et al., 2000; Macpherson et al., 2012). This may one of the reasons that the mortality rate is higher in the larval breeding stage. Applications of antibiotics do not effectively control the growth and reproduction of undesirable microbes, and easily lead to the emergence of antibiotic resistance of pathogenic bacteria (Smith et al., 1994; Muñoz-Atienza et al., 2014; Jiang et al., 2018). Consequently, the prevention and control of bacteriosis through biological method is gradually improvement and prevalent with the increasing awareness of food safety of people.
In this study, the black rockfish larvae and a juvenile were all healthy without any diseases although the abundances of these pathogenic species were higher, which suggested the environmental and/ or quantitative factors could not stimulate the relative diseases outbreak, and probiotics must not be absent in guts. These all indicated that the intestinal microbiota could maintain in a dynamic balance with the co-actions of pathogenic bacteria and potential probiotics. Meantime, the microbial quality of feed and water should be paid with great attention in larval breeding and cultured work.
5 CONCLUSIONThe intestinal microflora structure was gradually changed during the developmental stage of black rockfish larvae. However, the compositions of dominant species were always similar. Acidovorax, Aliivibrio, Bacillus, Acinetobacter, Pseudomonas, Lactobacillus, Lactococcus, Glaciecola, Vibrio, and Pseudoalteromonas are the core microbiota in this study. Through the relative analysis, we found Acidovorax, Glaciecola, Pseudomonas, Lactobacillus, and Acinetobacter might come from the parents of black rockfish larvae and/or fertilization process. Moreover, the effect of microbiota structure in feed on that in the fish gut was more obvious.
6 DATA AVAILABILITY STATEMENTThe datasets generated and analyzed during the current study available from the corresponding author on reasonable request.
Electronic supplementary material Supplementary material (Supplementary Tables S1–S4, Figs.S1–S2) is available in the online version of this article at https://doi.org/10.1007/s00343-019-9011-2.
Adhikari M, Yadav D R, Kim S W, Um Y H, Kim H S, Lee S C, Song J Y, Kim H G, Lee Y S. 2017. Biological control of bacterial fruit blotch of watermelon pathogen (Acidovorax citrulli) with rhizosphere associated bacteria. Plant Pathology Journal, 33(2): 170-183.
|
Alcaide E, Amaro C, Todoli R, Oltra R. 1999. Isolation and characterization of Vibrio parahaemolyticus causing infection in Iberian toothcarp Aphanius iberus. Diseases of Aquatic Organisms, 35(1): 77-80.
|
Austin B, Zhang X H. 2006. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Letters in Applied Microbiology, 43(2): 119-124.
DOI:10.1111/j.1472-765X.2006.01989.x |
Baik K S, Park Y D, Seong C N, Kim E M, Bae K S, Chun J. 2006. Glaciecola nitratireducens sp. nov., isolated from seawater. International Journal of Systematic and Evolutionary Microbiology, 56(9): 2 185-2 188.
DOI:10.1099/ijs.0.64330-0 |
Bakke I, Skjermo J, Vo T A, Vadstein O. 2013. Live feed is not a major determinant of the microbiota associated with cod larvae (Gadus morhua). Environmental Microbiology Reports, 5(4): 537-548.
DOI:10.1111/1758-2229.12042 |
Banerjee G, Ray A K. 2017. Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis, 72(1): 1-11.
DOI:10.1007/s13199-016-0441-8 |
Beurmann S, Ushijima B, Videau P, Svoboda C M, Smith A M, Rivers O S, Aeby G S, Callahan S M. 2017. Pseudoalteromonas piratica strain OCN003 is a coral pathogen that causes a switch from chronic to acute Montipora white syndrome in Montipora capitata. PLoS One, 12(11): e0188319.
DOI:10.1371/journal.pone.0188319 |
Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30(15): 2 114-2 120.
DOI:10.1093/bioinformatics/btu170 |
Chantharasophon K, Warong T, Mapatsa P, Leelavatcharamas V. 2011. High potential probiotic Bacillus species from gastro-intestinal tract of Nile Tilapia (Oreochromis niloticus). Biotechnology, 10(6): 498-505.
DOI:10.3923/biotech.2011.498.505 |
Chen J G, Yang J F, Xiong J, Mao Z J, Wangh L. 2010. The innate immune response in Lateolabrax japonicus induced by lipopolysaccharide from Glaciecola polaris strain ARK149 (LMG21854). Agricultural Sciences in China, 9(10): 1 504-1 511.
DOI:10.1016/S1671-2927(09)60245-5 |
Chun S J, Cui Y S, Ko S R, Lee H G, Srivastava A, Oh H M, Ahn C Y. 2017. Acidovorax lacteus sp. nov., isolated from a culture of a bloom-forming cyanobacterium (Microcystis sp.). Antonie Van Leeuwenhoek, 110(9): 1 199-1 205.
|
Chung H, Pamp S J, Hill J A, Surana N K, Edelman S M, Troy E B, Reading N C, Villablanca E J, Wang S, Mora J R, Umesaki Y, Mathis D, Benoist C, Relman D A, Kasper D L. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell, 149(7): 1 578-1 593.
DOI:10.1016/j.cell.2012.04.037 |
Dawood M A O, Koshio S. 2016. Recent advances in the role of probiotics and prebiotics in carp aquaculture: a review. Aquaculture, 454: 243-251.
DOI:10.1016/j.aquaculture.2015.12.033 |
Desai A R, Links M G, Collins S A, Mansfield G S, Drew M D, Van Kessel A G, Hill J E. 2012. Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhynchus mykiss). Aquaculture, 350-353: 134-142.
DOI:10.1016/j.aquaculture.2012.04.005 |
Edgar R C, Haas B J, Clemente J C, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16): 2 194-2 200.
DOI:10.1093/bioinformatics/btr381 |
Ferguson H W, Collins R O, Moore M, Coles M, MacPhee D D. 2004. Pseudomonas anguilliseptica infection in farmed cod, Gadus morhua L. Journal of Fish Disease, 27(4): 249-253.
DOI:10.1111/j.1365-2761.2004.00537.x |
Ferreira C, Veldhoen M. 2012. Host and microbes date exclusively. Cell, 149(7): 1 428-1 430.
DOI:10.1016/j.cell.2012.06.005 |
Forsythe P, Bienenstock J. 2010. Immunomodulation by commensal and probiotic bacteria. Immunological Investigations, 39(4-5): 429-448.
DOI:10.3109/08820131003667978 |
Gacias M, Gaspari S, Santos P M G, Tamburini S, Andrade M, Zhang F, Shen N, Tolstikov V, Kiebish M A, Dupree J L, Zachariou V, Clemente J C, Casaccia P. 2016. Microbiotadriven transcriptional changes in prefrontal cortex override genetic differences in social behavior. eLife, 5: e13442.
DOI:10.7554/eLife.13442 |
Gallo R L, Nakatsuji T. 2011. Microbial symbiosis with the innate immune defense system of the skin. Journal of Investigative Dermatology, 131(10): 1 974-1 980.
DOI:10.1038/jid.2011.182 |
Ghanbari M, Kneifel W, Domig K J. 2015. A new view of the fish gut microbiome: advances from next-generation sequencing. Aquaculture, 448: 464-475.
DOI:10.1016/j.aquaculture.2015.06.033 |
Hjerde E, Lorentzen M S, Holden M T G, Seeger K, Paulsen S, Bason N, Churcher C, Harris D, Norbertczak H, Quail M A, Sanders S, Thurston S, Parkhill J, Willassen N P, Thomson N R. 2008. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics, 9(1): 616.
DOI:10.1186/1471-2164-9-616 |
Horne M T, Baxendale A. 1983. The adhesion of Vibrio anguillarum to host tissues and its role in pathogenesis. Journal of Fish Diseases, 6(5): 461-471.
DOI:10.1111/j.1365-2761.1983.tb00100.x |
Jiang Y, Liu Z X, Liu X Z, Xu Y J, Shi B, Wang B. 2019. Structural characteristics and succession of intestinal microbiota for Paralichthys olivaceus during the early life stage. Aquaculture Research, 50(2): 529-540.
|
Jiang Y, Zhang Z, Wang Y G, Jing Y Y, Liao M J, Rong X J, Li B, Chen G P, Zhang H S. 2018. Effects of probiotic on microfloral structure of live feed used in larval breeding of turbot Scophthalmus maximus. Journal of Oceanology and Limnology, 36(3): 1 002-1 012.
DOI:10.1007/s00343-018-7049-1 |
Karasov W H, Martínez del Rio C, Caviedes-Vidal E. 2011. Ecological physiology of diet and digestive systems. Annual Review of Physiology, 73: 69-93.
DOI:10.1146/annurev-physiol-012110-142152 |
Karlsen C, Vanberg C, Mikkelsen H, Sørum H. 2014. Coinfection of Atlantic salmon (Salmo salar), by Moritella viscosa and Aliivibrio wodanis, development of disease and host colonization. Veterinary Microbiology, 171(1-2): 112-121.
DOI:10.1016/j.vetmic.2014.03.011 |
Kelly P. 2010. Nutrition, intestinal defence and the microbiome. Proceedings of the Nutrition Society, 69(2): 261-268.
DOI:10.1017/S0029665110000108 |
Kim Y R, Kim E Y, Choi S Y, Hossain M T, Oh R K, Heo W S, Lee J M, Cho Y C, Kong I S. 2013. Effect of a probiotic strain, Enterococcus faecium, on the immune responses of olive flounder (Paralichthys olivaceus). Journal of Microbiology and Biotechnology, 22(4): 526-529.
|
Kitani Y, Kikuchi N, Zhang G H, Ishizaki S, Shimakura K, Shiomi K, Nagashima Y. 2008. Antibacterial action of L-amino acid oxidase from the skin mucus of rockfish Sebastes schlegelii. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 149(2): 394-400.
DOI:10.1016/j.cbpb.2007.10.013 |
Kozińska A, Paździor E, Pękala A, Niemczuk W. 2014. Acinetobacter johnsonii and Acinetobacter lwoffii-the emerging fish pathogens. Bulletin of the Veterinary Institute in Pulawy, 58(2): 193-199.
DOI:10.2478/bvip-2014-0029 |
Leyton Y, Sayes C, Mejias C, Abarca M, Wilson R, Riquelme C. 2017. Increased larval survival of Seriola lalandi using Pseudoalteromonas sp. as probiotics. Revista de Biología Marina Y Oceanografía, 52(1): 95-101.
DOI:10.4067/S0718-19572017000100007 |
Li J, Cao J L, Wang X, Liu N, Wang W M, Luo Y. 2017a. Acinetobacter pittii, an emerging new multi-drug resistant fish pathogen isolated from diseased blunt snout bream (Megalobrama amblycephala Yih) in China. Applied Microbiology and Biotechnology, 101(16): 6 459-6 471.
DOI:10.1007/s00253-017-8392-4 |
Li X M, Yu Y H, Feng W S, Yan Q Y, Gong Y C. 2012. Host species as a strong determinant of the intestinal microbiota of fish larvae. Journal of Microbiology, 50(1): 29-37.
|
Li Y X, Yang P, Zhang Y J, Ai Q H, Xu W, Zhang W B, Zhang Y A, Hu H B, Liu J T, Mai K S. 2017b. Effects of dietary glycinin on the growth performance, digestion, intestinal morphology and bacterial community of juvenile turbot, Scophthalmus maximus L. Aquaculture, 479: 125-133.
DOI:10.1016/j.aquaculture.2017.05.008 |
Liu P C, Lin J Y, Hsiao P T, Lee K K. 2004. Isolation and characterization of pathogenic Vibrio alginolyticus from diseased cobia Rachycentron canadum. Journal of Basic Microbiology, 44(1): 23-28.
DOI:10.1002/jobm.200310316 |
Liu Y, Li N Q, Zhao X P, Yue B, He S W, Gao Z X, Zhou S, Zhang M. 2016. A C-type lectin that inhibits bacterial infection and facilitates viral invasion in black rockfish, Sebastes schlegelii. Fish & Shellfish Immunology, 57: 309-317.
|
López-Romalde S, Magariños B, Nuñez S, Toranzo A E, Romalde J L. 2003. Phenotypic and genetic characterization of Pseudomonas anguilliseptica strains isolated from fish. Journal of Aquatic Animal Health, 15(1): 39-47.
DOI:10.1577/1548-8667(2003)015<0039:PAGCOP>2.0.CO;2 |
Lynch III J P, Zhanel G G, Clark N M. 2017. Infections due to Acinetobacter baumannii in the ICU: treatment Options. Seminars in Respiratory and Critical Care Medicine, 38(3): 311-325.
DOI:10.1055/s-0037-1599225 |
Macpherson H L, Bergh Ø, Birkbeck T H. 2012. An aerolysinlike enterotoxin from Vibrio splendidus may be involved in intestinal tract damage and mortalities in turbot, Scophthalmus maximus (L.), and cod, Gadus morhua L., larvae. Journal of Fish Diseases, 35(2): 153-167.
|
Magi G E, Lopez-Romalde S, Magariños B, Lamas J, Toranzo A E, Romalde J L. 2009. Experimental Pseudomonas anguilliseptica infection in turbot Psetta maxima (L.): a histopathological and immunohistochemical study. European Journal of Histochemistry, 53(2): e9.
DOI:10.4081/ejh.2009.e9 |
Muñoz-Atienza E, Araújo C, Magadán S, Hernández P E, Herranz C, Santos Y, Cintas L M. 2014. In vitro and in vivo evaluation of lactic acid bacteria of aquatic origin as probiotics for turbot (Scophthalmus maximus L.) farming. Fish & Shellfish Immunology, 41(2): 570-580.
|
Nicholson J K, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science, 336(6086): 1 262-1 267.
DOI:10.1126/science.1223813 |
Nossa C W, Oberdorf W E, Yang L Y, Aas J A, Paster B J, DeSantis T Z, Brodie E L, Malamud D, Poles M A, Pei Z H. 2010. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World Journal of Gastroenterology, 16(33): 4 135-4 144.
DOI:10.3748/wjg.v16.i33.4135 |
Ntranos A, Casaccia P. 2018. The Microbiome-gut-behavior axis: crosstalk between the gut microbiome and oligodendrocytes modulates behavioral responses. Neurotherapeutics, 15(1): 31-35.
DOI:10.1007/s13311-017-0597-9 |
Pérez T, Balcázar J L, Ruiz-Zarzuela I, Halaihel N, Vendrell D, de Blas I, Múzquiz J L. 2010. Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunology, 3(4): 355-360.
DOI:10.1038/mi.2010.12 |
Pérez-Sánchez T, Ruiz-Zarzuela I, de Blas I, Balcázar J L. 2014. Probiotics in aquaculture: a current assessment. Reviews in Aquaculture, 6(3): 133-146.
DOI:10.1111/raq.12033 |
Qian G Y, Xu Z R. 2007. Effect of polysaccharide extracted from Glaciecola polaris on the protection of mouse macrophages from oxidative injury. Bioresource Technology, 98(1): 202-206.
DOI:10.1016/j.biortech.2005.12.004 |
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner F O. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research, 41(D1): D590-D596.
|
Reddy M R K, Mastan S A. 2013. Emerging Acinetobacter schindleri in red eye infection of Pangasius sutchi. African Journal of Biotechnology, 12(50): 6 992-6 996.
|
Reyon D, Tsai S Q, Khayter C, Foden J A, Sander J D, Joung J K. 2012. FLASH assembly of TALENs for high-throughput genome editing. Nature Biotechnology, 30(5): 460-465.
DOI:10.1038/nbt.2170 |
Richards G P, Watson M A, Needleman D S, Uknalis J, Boyd E F, Fay J P. 2017. Mechanisms for Pseudoalteromonas piscicida-induced killing of vibrios and other bacterial pathogens. Applied and Environmental Microbiology, 83(11): e00175.
DOI:10.1128/AEM.00175-17 |
Ringø E, Olsen R E, Mayhew T M, Myklebust R. 2003. Electron microscopy of the intestinal microflora of fish. Aquaculture, 227(1-4): 395-415.
DOI:10.1016/j.aquaculture.2003.05.001 |
Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ, 4: e2584.
DOI:10.7717/peerj.2584 |
Sanchez L M, Wong W R, Riener R M, Schulze C J, Linington R G. 2012. Examining the fish microbiome: vertebratederived bacteria as an environmental niche for the discovery of unique marine natural products. PLoS One, 7(5): e35398.
DOI:10.1371/journal.pone.0035398 |
Shabat S K B, Sasson G, Doron-faigenboim A, Durman T, Yaacoby S, Berg Miller M E, White B A, Shterzer N, Mizrahi I. 2016. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. The ISME Journal, 10(12): 2 958-2 972.
DOI:10.1038/ismej.2016.62 |
Smith P, Hiney M P, Samuelsen O B. 1994. Bacterial resistance to antimicrobial agents used in fish farming: a critical evaluation of method and meaning. Annual Review of Fish Diseases, 4: 273-313.
DOI:10.1016/0959-8030(94)90032-9 |
Sullam K E, Essinger S D, Lozupone C A, O'Connor M P, Rosen G L, Knight R, Kilham S S, Russell J A. 2012. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Molecular Ecology, 21(13): 3 363-3 378.
DOI:10.1111/j.1365-294X.2012.05552.x |
Swain P, Nayak S K, Nanda P K, Dash S. 2008. Biological effects of bacterial lipopolysaccharide (endotoxin) in fish: a review. Fish & Shellfish Immunology, 25(3): 191-201.
|
Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, Shirakawa T, Sonomoto K, Nakayama J. 2009. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunology & Medical Microbiology, 56(1): 80-87.
|
Urdaci M C, Bressollier P, Pinchuk I. 2004. Bacillus clausii probiotic strains: antimicrobial and immunomodulatory activities. Journal of Clinical Gastroenterology, 38(S6): S86-S90.
|
Verschuere L, Rombaut G, Sorgeloos P, Verstraete W. 2000. Probiotic bacteria as biological control agents in aquaculture. Microbiology and Molecular Biology Reviews, 64(4): 655-671.
DOI:10.1128/MMBR.64.4.655-671.2000 |
Wang Q, Garrity G M, Tiedje J M, Cole J R. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73(16): 5 261-5 267.
DOI:10.1128/AEM.00062-07 |
Wong D, Nielsen T B, Bonomo R A, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clinical Microbiology Reviews, 30(1): 409-447.
|
Yan L C, Hu B S, Chen G, Zhao M, Walcott R R. 2017. Further evidence of cucurbit host specificity among Acidovorax citrulli groups based on a detached melon fruit pathogenicity assay. Phytopathology, 107(11): 1 305-1 311.
DOI:10.1094/PHYTO-11-16-0416-R |
Ye L, Amberg J, Chapman D, Gaikowski M, Liu W T. 2014. Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. The ISME Journal, 8(3): 541-551.
DOI:10.1038/ismej.2013.181 |
Zhang Z G, Xu D M, Wang L, Hao J J, Wang J F, Zhou X, Wang W W, Qiu Q, Huang X D, Zhou J W, Long R J, Zhao F Q, Shi P. 2016. Convergent evolution of rumen microbiomes in high-altitude mammals. Current Biology, 26(14): 1 873-1 879.
DOI:10.1016/j.cub.2016.05.012 |
Zhao Y C, Yuan L, Wan J L, Sun Z X, Wang Y Y, Sun H S. 2016. Effects of potential probiotic Bacillus cereus EN25 on growth, immunity and disease resistance of juvenile sea cucumber Apostichopus japonicus. Fish & Shellfish Immunology, 49: 237-242.
|
Ziaei-Nejad S, Rezaei M H, Takami G A, Lovett D L, Mirvaghefi A R, Shakouri M. 2006. The effect of Bacillus spp. bacteria used as probiotics on digestive enzyme activity, survival and growth in the Indian white shrimp Fenneropenaeus indicus. Aquaculture, 252(2-4): 516-524.
DOI:10.1016/j.aquaculture.2005.07.021 |
Zorrilla I, Arijo S, Chabrillon M, Diaz P, Martinez-Manzanares E, Balebona M C, Moriñigo M A. 2003. Vibrio species isolated from diseased farmed sole, Solea senegalensis (Kaup), and evaluation of the potential virulence role of their extracellular products. Journal of Fish Diseases, 26(2): 103-108.
DOI:10.1046/j.1365-2761.2003.00437.x |
 2020, Vol. 38
2020, Vol. 38