Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CUI Wenxiao, MA Aijun, HUANG Zhihui, WANG Xin'an, SUN Zhibin, LIU Zhifeng, ZHAN GWei, YANG Jingkun, ZHANG Jinsheng, QU Jiangbo
- Transcriptomic analysis reveals putative osmoregulation mechanisms in the kidney of euryhaline turbot Scophthalmus maximus responded to hypo-saline seawater
- Journal of Oceanology and Limnology, 38(2): 467-479
- http://dx.doi.org/10.1007/s00343-019-9056-2
Article History
- Received Mar. 8, 2019
- accepted in principle Jul. 10, 2019
- accepted for publication Jul. 22, 2019
2 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266071, China;
3 Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences; Shandong Key Laboratory of Marine Fisheries Biotechnology and Genetic Breeding; Qingdao Key Laboratory for Marine Fish Breeding and Biotechnology, Qingdao 266071, China;
4 Yantai Tianyuan Aquatic Limited Corporation, Yantai 264003, China
As an inherent physicochemical property of water, salinity is one of the main factors limiting species distribution (Brennan et al., 2015). Euryhaline fish possess particular osmotic regulation mechanisms that enable them to sense and compensate for salinity fluctuation in the marine environment (Edwards and Marshall, 2012). They can perceive osmosensory signaling that activate control system, then coordinate effectors to counteract osmotic stress (Fiol and Kültz, 2007). The adaptability of euryhaline fish to the salinity in a dramatically changing aquatic environment is essential for survival, and provides a classical model for investigating osmotic regulation (Lam et al., 2014). The osmoregulatory mechanisms that euryhaline fish harbor include complex functional networks encoded by widely distributed functional proteins (Kültz, 2015). Using transcriptomics approaches, it was shown that the functional distributions of differentially expressed genes (DEGs) found in the kidney is wide than in the gill of the European eel (Anguilla anguilla) (Kalujnaia et al., 2007).
Turbot (Scophthalmus maximus) is a euryhaline flounder with great commercial value that is native to the North Atlantic and its saltwater-freshwater border (Pereiro et al., 2012). Turbot have a relatively remarkable ability to adapt to opposing osmotic challenges and have been proven an excellent model species to study physiological adaptations of flounder associated with osmoregulatory plasticity. There have been many studies on the adaptability of turbot to salinity mainly in the aspects of physiology (Imsland et al., 2001; Tang et al., 2006; Dietz et al., 2013), but the osmoregulatory mechanism at the transcriptional level is still unknown. Osmoregulation in teleosts is mainly mediated by the kidneys, gills, and intestine (Marshall and Grosell, 2005). Turbot kidney is an important organ of osmotic regulation, which plays an irreplaceable role in osmotic regulation (Prodocimo et al., 2008). Kidney filtration and reabsorption is important for osmoregulation as any excess uptake of divalent ions is excreted by the kidney (Beyenbach, 1986, 2004). Moreover, some osmoregulatory hormones in teleost fishes, such as renin and cortisol, are secreted by the kidney and are critical for body fluid regulation through its effect on tubular water reabsorption in the kidney (Babey et al., 2011). In addition, there are a large number of special osmoregulatory effectors, osmotic stress signaling and ion channels acting on osmoregulation in the kidney (Kasprowicz, 2011).
In this study, we employed next-generation sequencing (NGS) technology to sequence and assemble the de novo kidney transcriptome of euryhaline turbot in response to a hypo-osmotic environment. First, the transcripts were assembled to unigenes the Trinity transcriptome assembler. Subsequently, GO and KEGG enrichment analyses revealed unigenes that are involved in different biological process and signaling pathways. The aims of this study were to identify transduction processes of osmotic pressure signaling and effector mechanism response to low salinity stress. Although variations in transcript abundance does not mean the variations of protein contents, nevertheless, this study identified many crucial biological processes and signaling pathways involved in osmoregulation from the perspective of transcriptome in kidneys under low salinity stress. To the best of our knowledge, this was the first NGS study to provide a transcriptome-wide perspective on the mechanisms of osmoregulation in turbot, especially the process of signal transduction in kidneys of euryhaline fish in response to hyposalinity.
2 MATERIAL AND METHOD 2.1 Material 2.1.1 Ethics statementThe handing of turbot was performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The study procedures were approved by the Experimental Animal Ethics Committee in Chinese Academy of Fishery Sciences, China.
2.1.2 FishThe healthy juvenile turbot (12.52±0.34 cm) were from Yantai Tianyuan Aquaculture Co., Ltd. in Shandong Province, China. Healthy fish were maintained for 2 weeks in 150-L tanks. Water was aerated to keep normoxic oxygen saturation at all times and the water was not exchanged. Diluted seawater (salinity of 5, 10) was obtained by mixing freshwater with seawater (salinity of 30) taken from a deep well, whereas hypersaline water (salinity of 50) was obtained by mixing seawater crystals with seawater. We sampled intestine, kidney, gill, liver, skin, spleen, and brain tissues from turbot that had been treated in their respective diluted seawater or seawater, and hypersaline environments for 24 h. Before sampling, fish individuals were anesthetized with MS-222. Three biological replicates were set in separate aquarium with same aquatic condition in each experiment. The fresh tissues were immersed in RNAstor immediately at 4℃ overnight and then frozen at -80℃ until the next work.
2.2 Method 2.2.1 Transcriptome sequencingTotal RNA was extracted from kidney tissue (salinity of 5 and 30) samples (n=3) using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and RNA quality and quantity were evaluated in a Qubit and a Nanodrop, respectively. After checking RNA purity and concentration, the integrity of RNA samples was assessed using an Agilent 2100 instrument (Agilent, Santa Clara, CA, USA). RNAs from individual fish in each group were used to construct cDNA libraries. The six cDNA libraries were constructed and subjected to pair-end sequencing on an Illumina Hiseq 2000 platform at Novogene Bioinformatics Technology Co. Ltd. (Beijing, China).
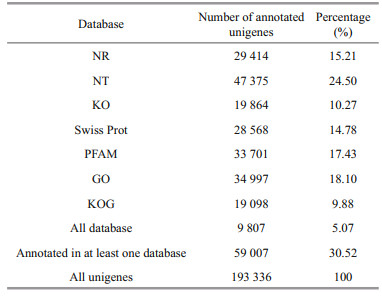
2.2.2 Quality control, assembly, and gene annotationRaw reads were produced through base calling and stored in fastq format. The raw data were cleaned after filtration by removing the adaptors, poly-N of more than 10%, and low-quality sequences (base quality score < Q20). Transcriptome assembly was performed using Trinity software (Grabherr et al., 2011) as described for de novo transcriptome assembly without the reference genome. The k-mer value was set to 25 and min_kmer_cov set to 2, by default. The longest assembled sequences of each transcription set were defined as unigenes. Genes encoding protein domains were identified by searching against non-redundant (NR), NT, Swiss-Prot, PFAM databases through the BLASTp. GO functions for all unigenes were classified using Gene2go v2.5 (Götz et al., 2008) with an E-value threshold of 1e-6. The pathway annotations and classifications of unigenes were performed based on KEGG and COG databases using the BLAST, with an E-value threshold of 1e-10 and 1e-3, respectively.
Trinity was used to assemble clean reads into the sequences by RSEM (Li and Dewey, 2011). The expression of genes was normalized and calculated by the FPKM (Fragments Per kb per Million fragments) method (Trapnell et al., 2010). Differential expression of unigenes was analyzed using the MAplot-based method with random sampling model (MARS) in DEGseq (Anders and Huber, 2010). The P value was adjusted by means of the q value. A padj < 0.05 was set as the threshold for significantly differential expression, according to biological repetition. Functional categories analysis on Go database of the DEGs was performed using GOseq (Young et al., 2010) based on a hypergeometric distribution test. GO terms with false discovery rates (FDR) ≤0.05 were defined as significantly enriched in DEGs. Using KOBAS (2.0) software, pathway enrichment analysis of the DEGs based on KEGG was performed (Mao et al., 2005).
2.2.3 Fluorescence quantitative PCR and statistical analysisTotal RNA was extracted from tissues treated with four gradients of salinity using an RNA prep pure Tissue Kit (Tiangen, Beijing, China). First strand cDNA was synthesized by reverse transcription using Quant Reverse Transcriptase. All primers for qPCR were designed using Primer5.0 software. Primers are listed in Supplementary Table S1. Real-time PCR was performed on a 185-5096 CFX96 Real-Time PCR Detection System (Beijing yue da Biotechnology Co. Ltd., Beijing, China) in a 15-μL reaction using 1 μL cDNA, 10 μL 2×TransStart Top Green qPCR SuperMix, 0.4 μL forward and reverse primers, 7.8 μL RNase-Free ddH2O and 0.4 μL 50×Passive Reference Dye 1. Cycling conditions were 94℃ for 30 s followed by 40 cycles of 95℃ for 5 s, and 60℃ for 30 s. The blastN and blastX tools were used for a homogeneity search available online at http://blast.ncbi.nlm.nih.gov/Blast. All quantitative data shown are the means ± S.E.M. of multiple samples (n=3). The expression of genes was analyzed by the 2-ΔΔCt method (Schmittgen and Zakrajsek, 2000). Statistics software SPSS19.0 (SPSS, Chicago, IL, USA) was used to test significance. The significance threshold was set at P < 0.05 for all tests.
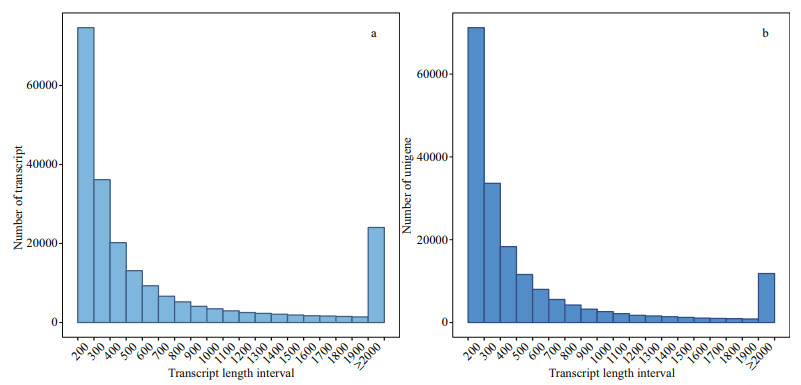
3 RESULT AND DISCUSSION 3.1 Sequencing and de novo assembly of kidney transcriptomesThe results showed that in total an average of 42 652 674 clean reads were obtained from the six libraries of turbot samples under salinity stress. 193 336 unigenes were obtained from 226 176 transcripts assembled with clean reads (Fig. 1). As shown, the length distribution of all unigenes ranged from 201 to 25 839 bp, with an average length of 786 bp.
|
| Fig.1 Summary statistics of sequencing output and de novo assembly of kidney transcriptome a. transcript length distribution; b. unigene length distribution. |
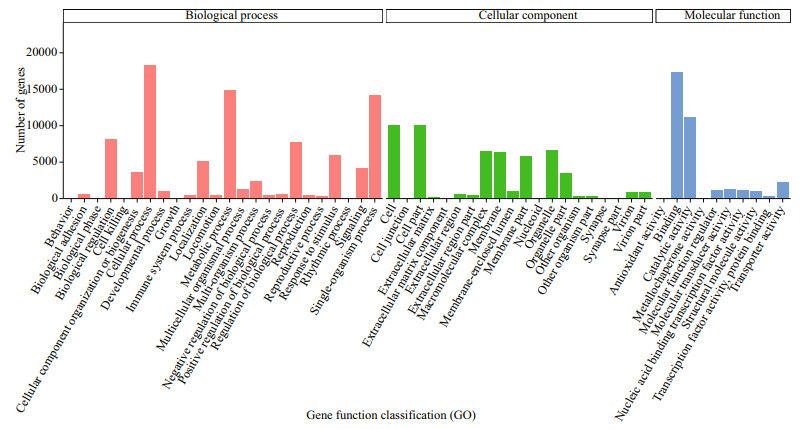
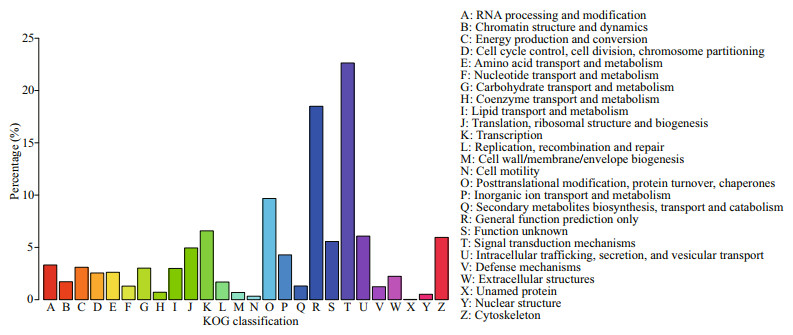
In order to determine their function, all unigenes were blast analyzed against publicly available databases. All unigenes had significant hits in the NCBI NR, NT, PFAM, Swiss Prot, KOG, GO, and KEGG databases (Table 1). Overall, 59 007 unigenes (30.52%) were annotated in at least one database, and 47 375 (24.50%), and 19 098 (9.88%) unigenes were annotated in the NT and KOG databases, respectively. According to the results annotated in the GO database, 34 997 unigenes were divided into 54 subcategories (Fig. 2). A total of 19 098 unigenes were annotated into 26 different processes on the KOG database, and the largest category was represented by "signal transduction mechanisms", which was followed by "general function prediction only", and "posttranslational modification, protein turnover, chaperones" (Fig. 3). The total annotated unigenes were classified into five major functional categories: cellular processes, environmental information processing, genetic information processing, metabolism, and organismal systems, which were further divided into 4, 3, 4, 12, and 9 subcategories, respectively (Fig. 4), and the largest category was represented by "signal transduction", which was followed by "endocrine system".
|
| Fig.2 Categories of GO |
|
| Fig.3 Categories of KOG |

|
| Fig.4 Categories of KEGG A. cellular processes; B. environmental information processing; C. genetic information processing; D. metabolism; E. organismal systems. |
To assess the DEGs obtained by RNA sequencing for the reliability of transcriptional data, qPCR was carried out with the total RNA used for RNA sequencing, to examine the expression of 10 DEGs in kidneys under salinities of 5 and 30, respectively, which included six upregulated unigenes and four downregulated unigenes (Fig. 5a). Among the 10 selected genes, the relative expression of seven genes between brackish water- and seawater-treated kidneys matched with transcriptomes data, except genes APOM, AATP6V1E1 and H4. Our results showed that expression levels of the ten genes were obviously changed after salinity stress. This suggested that the real-time PCR data for hypo-salinity treated kidneys were in good concordance with the transcriptome data (Fig. 5b).
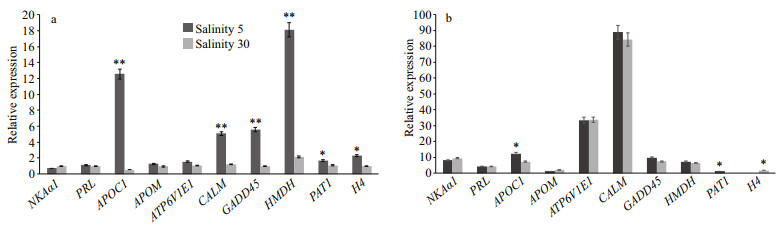
|
| Fig.5 Expression of 10 genes in kidney One asterisk (*) indicates P < 0.05, two asterisks (**) indicate P < 0.01. NKAα: Na+/K+-ATPase; PRL: prolactin; APOC1: apolipoprotein C-I; APOM: apolipoprotein M; ATP6V1E1: V-type proton ATPase subunit E 1; CALM: calmodulin; GADD45: Growth arrest and DNA-damage-inducible protein GADD45; HMDH: 3-hydroxy-3-methylglutaryl-coenzyme A reductase; PAT1: solute carrier family 26 (sulfate anion transporter): member 6; H4: histone H4. |
Analysis of the biological functions of DEGs showed that 23 significant enrichment pathways were upregulated and 16 of these were in the immunological category (Supplementary Table S2). There were 41 significant enrichment pathways in which genes were downregulated, other than legionellosis; these belonged to energetic metabolism related to digestion and absorption (Supplementary Table S3). We presumed that the immune system functions in hypoosmotic stress to defend turbot against changes of diversified external environmental factors involving in environmental salinity, temperature and autochthonous microorganisms (Narnaware et al., 2000). However, the mRNA expressions of genes in energetic metabolism pathways were reduced, which suggest SW acclimation was associated with decreased respiratory activity and energy production in kidney (Kalujnaia et al., 2007).
In hypo-osmotic brackish water, turbot need to reduce salt loss and water gain with kidney (Farrell, 2011). To maintain normal physiological functions, the signaling pathways related to cell morphology and cell volume regulation are upregulated (Lam et al., 2014). Moreover, the signaling pathways associated with maintaining cell morphology, by increasing ionocytes with the capacity to reabsorb dissolved salt lost to the filtrate, were upregulated in hypo-osmotic stress. Consequently, signaling pathways and biological processes that were enriched in the kidney involved apoptosis, the cell cycle, oocyte meiosis, the p53 signaling pathway, regulation of actin cytoskeleton, adherens junction, focal adhesion, gap junction, signaling pathways regulating pluripotency of stem cells, and tight junctions (Supplementary Table S4). These signaling pathways and biological processes could play important roles in cell growth and death, movement, and cellular interactions in response to osmotic stress, which has been documented in other cell types (Kalujnaia et al., 2007; Hoffmann et al., 2009; Lam et al., 2014). In agreement with previous studies (Kültz and Burg, 1998; Kültz and Avila, 2001), our analysis indicated that transcripts associated with MAPK signaling, an important pathway in the osmotic stress response, were abundant in the hypo-salinity treated kidney of turbot.
3.5 Analysis of signal transduction in hypo-osmotic regulationA total of 45 signal pathways were detected in 246 physiological processes of turbot hypoosmoregulation, of which 25 signal pathways belonged to the category of signal transduction (Supplementary Table S5). Differential expression analysis showed that the expression profiles of the genes encoding core protein, as well as its upstream ligands and downstream acting protein genes, of nine out of 45 signaling pathways, changed. The results revealed that the ErbB, FoxO, Jak-STAT, MAPK, Notch, PI3K-Akt, phosphatidylinositol, Ras, and cGMP-PKG signaling pathway, which belong to the category of signal transduction, may play an important role in signal transduction of ionic and osmotic regulation, and the control of extracellular fluid volume in the kidney of turbot. Among these signal pathways, the ErbB, FoxO, Jak-STAT, MAPK, Notch, Ras, and cGMP-PKG signaling pathway belong to signal transduction mediated by enzyme-coupled receptors, while the PI3K-Akt signaling pathway and phosphatidylinositol signaling pathways are mediated by G-protein coupled receptors (Bonni et al., 1997; Vojtek and Der, 1998; Grant et al., 2002; Arden, 2004; Ehebauer et al., 2006). However, signal transduction using an enzyme- coupled receptor and G-protein coupled receptor require that the extracellular ligand (signal molecule) binds to the cell receptor to form a complex that acts on the target protein (Bakir and Sezerman, 2006).
Interplay between signaling pathways (Fig. 6) indicated that the MAPK signaling pathway participated in the biological function of seven signaling pathways other than the phosphatidylinositol signaling pathway, and the PI3K-Akt signaling pathway interacted with five signaling pathways except for the phosphatidylinositol signaling pathway, Notch signaling pathway, and MAPK signaling pathway. Therefore, the importance of the MAPK signaling pathway (Fouchs et al., 2010) and PI3K-Akt signaling pathway (Xu et al., 2015) in the osmotic adjustment of turbot in a hypo-osmotic environment was also demonstrated, and also showed that two signaling pathways may integrate and precisely control multiple signals in cells to form a signaling network system and finally control the appropriate response.
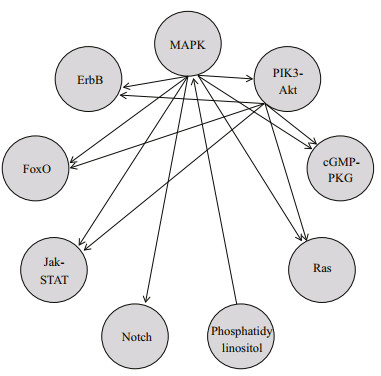
|
| Fig.6 Interaction of nine signaling pathways A→B: A functioned in B. |
Analysis of the ligands of biological processes and signaling pathways enriched in the endocrine system suggested that the physiological productions of biological processes were involved in insulin secretion, melanogenesis, renin secretion, renin-angiotensin system, thyroid hormone synthesis, and signaling molecules of signaling pathways involving glucagon signaling pathway, GnRH signaling pathway, insulin signaling pathway, oxytocin signaling pathway, PPAR signaling pathway, prolactin signaling pathway, and the thyroid hormone signaling pathway, and all were through proteins. In addition, the results of DEGs showed that only the mRNA of genes encoding core proteins of renin secretion had changed. The analysis showed that renin was likely to be generated in hypoosmoregulation. Our findings corroborated those of a previous report on the renin-angiotensin-aldosterone system, which occupy a crucial position in maintaining of homeostasis electrolyte and fluid, and in enhancing reabsorption of Na+ from kidney (Kurtz A, Wagner, 1999). Signal transduction in the organism requires interactions between the signal molecule and the receptor (Bhattacharya, 2010). In the signal transduction process of hypo-osmoregulation in turbot, biological processes that were enriched in signaling molecules and interactions involved cell adhesion molecules (CAMs), cytokine-cytokine receptor interactions, extracellular matrix (ECM)- receptor interactions, and neuroactive ligand-receptor interactions. The analysis of the amount and abundance of genes expressed in physiological processes suggested the importance of ECM-receptor interaction, cytokine-cytokine receptor interaction, neuroactive ligand-receptor interaction, and CAMs in signal transduction decreased in hypo-osmoregulation.
Cell signal transduction is a key process of cellto-cell communication essential for the coordination of cell activities, the control of cell growth and division, generation of tissues, and formation of morphology in multicellular organisms (Rappolee and Armant, 2009). Cells can communicate with each other not only by secreting chemical signals but also by cell contact and gap junction signaling (Kurt et al., 2011). The analysis of the enrichment of biological processes indicated that tight junction, adherens junction, focal adhesion, gap junction, signaling pathways regulating pluripotency of stem cells, were enriched in the cellular community. In addition, according to the mode of action, all played a role in cell-cell junctions, while focal adhesion also acted on the connection of cells to the ECM; however, all of the linkages acted on transmembrane transport and signal transduction. Analyses of the DEGs and enrichment of biological processes revealed that the mRNA of genes encoding the core proteins of adherens junctions and focal adhesion, important biological processes in the hypo-osmotic stress response, had changed. Therefore, the gap junctions that exist widely in animal tissues (Shibata et al., 2001) may play an important role in hypoosmoregulation signal transduction in turbot.
The conduction of nerve impulses is completed through the synapses in neurons-neurons or neuronseffector cells (such as muscle cells). It was found that signaling pathways involving cholinergic synapse, dopaminergic synapse, GABAergic synapse, glutamatergic synapse, and serotonergic synapse were enriched in the nervous system. More importantly, all synapses are chemical. However, chemical synapses are biological junctions through which the signals of neurons can be exchanged with each other and with non-neuronal cells such as those in muscles or glands, whereas electrical synapses directly transfer electrical signals from one cell to another through gap junctions. Therefore, gap junctions associated with electrical synaptic signaling may not play a significant role in hypo-osmoregulation signal transduction in turbot. Analysis of the genes of chemical post-synaptic cell receptors and receptorrelated proteins showed differential expressions of genes associated with neurotransmitter receptors and receptor-related proteins of the GABAergic synapse, among the five chemical synapses. The differential expression underscored the importance of GABAergic synapses in hypo-osmoregulation signal transduction in turbot.
In summary, on the transcriptional level, biological processes involved in renin secretion, ECM-receptor interaction, adherens junction, and focal adhesion play an important role in the plasticity phenotype in hypo-stress, while the signal transduction network composed of the MAPK signaling pathway and PI3KAkt signaling pathway with the GABAergic synapse work in hypo-osmoregulation signal transduction in turbot.
3.6 Analysis of biological processes associated with inorganic channels and transportersThere are some studies on osmoregulation mechanisms of euryhaline fish kidney, and basic schemas involving in ion channels and transporters for salinity stress responses have been proposed (Kültz, 2015). By analyzing the roles of signal pathways and biological processes that were significantly enriched in genes, biological processes involving mineral absorption (Fig. 7) and bile secretion (Fig. 8) associated with iono-osmoregulation were identified. Several homologs of key ion channels and transporters that aid in absorbing salt, such as SLC5A1 (SGLT1), SLC9A3 (NHE-3), and SLC26A6 (PAT1), were identified as downregulated in brackishtreated kidney on mineral absorption, which confirmed previous reports (Inokuchi et al., 2008; Hummel et al., 2011; Karaica et al., 2015). Genes that were related to water molecule secretion, AQP1(aquaporin), AQP4, AQP8, and the genes related to Na+ uptake, ASBT (SLC10A2), SLC5A1 (SGLT1), and SLC9A3 (NHE-3), were down-regulated among 12 DEGs during the physiological process of bile secretion. The potential roles for aquaporin isoforms in water or solute transport in the gill of other fish have been discussed (Cutler and Cramb, 2002; Nishimoto et al., 2007; Cutler et al., 2009). In agreement with previous studies observed in higher vertebrates, SLC10A2 underscores the role of the ileal Na+/bile acid cotransporter in the intestinal reclamation of bile acids (Oelkers et al., 1997). However, the mRNA of genes NPT2b, B0AT1 (SLC6A19), NHE1 (SLC9A1), and NBC (SLC4A5) related to Na+ uptake were independent of the two physiological processes, presumably due to the diversity of gene function.
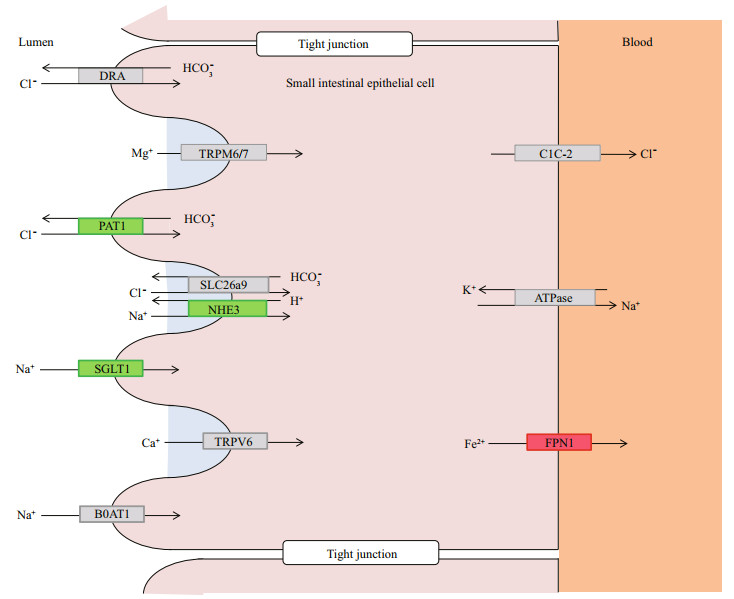
|
| Fig.7 Schematic view of inorganic ion channels and transporters involved in mineral absorption encoded by genes identified as differentially expressed in the kidney of turbot The downregulated genes are represented in green and upregulated genes in red while the none-differentially expressed genes are in gray. DRA (SLC26A3): solute carrier family 26 (chloride anion exchanger), member 3; TRPM6/7: transient receptor potential cation channel subfamily M member 6; PAT1 (SLC26A6): solute carrier family 26 (sulfate anion transporter), member 6; SLC26A9: solute carrier family 26 (sulfate anion transporter), member 9; NHE3 (SLC9A3): solute carrier family 9 (sodium/hydrogen exchanger), member 3; SGLT1 (SLC5A1): solute carrier family 5 (sodium/glucose cotransporter), member 1; TRPV6: transient receptor potential cation channel subfamily V member 6; B0AT1 (SLC6A19): solute carrier family 6 (neurotransmitter transporter) member 19; ClC-2 (CLCN2): chloride channel 2; ATPase: sodium/potassium-transporting ATPase subunit alpha; FPN1(SLC40A1): solute carrier family 40 (iron-regulated transporter), member 1. |
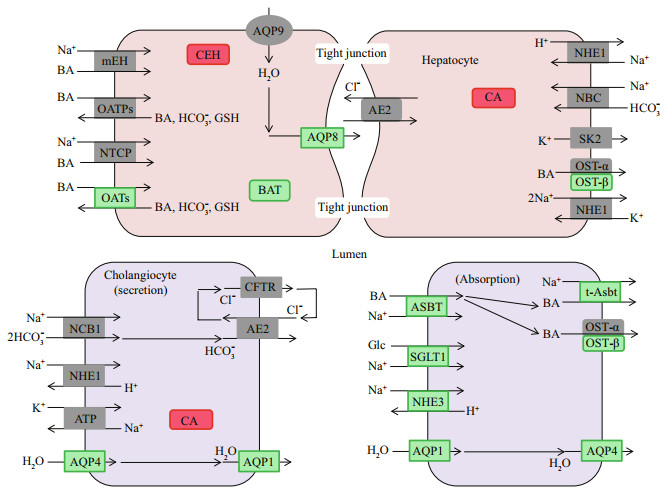
|
| Fig.8 Schematic view of inorganic ion channels and transporters involved in bile secretion encoded by genes identified as differentially expressed in the kidney of turbot The downregulated genes are represented in green and upregulated genes in red while the none-differentially expressed genes are in gray. |
We performed expression analyses using qPCR on crucial genes associated with biological processes or pathways functioning in hypo-osmotic acclimation. The relative expression levels as determined by realtime quantitative PCR are summarized in Fig. 9. Our results exhibited slightly similar expression patterns in response to salt stress. The lowest expression level of six genes was found in intestinal tissue in which the transcripts of the six genes were accumulated at the highest quantity at salinity 30 (control group). This indicates that the intestinal tissue was vulnerable to osmotic pressure and may play a role in osmoregulation in response to salt stress. The expression trend of gene NKAα1 first increased and then decreased, and was up-regulated to the highest level under salinity-10 treatment in all tissues except for intestinal tissue. The expression patterns of genes were different between tissues and salinity and suggested the specificity of the gene expression with organization and salinity. The highest expression level of gene PRL was found in spleen, followed by kidney and liver, while the highest expression level of gene PAT1 was found in the gill. As a freshwateradapting hormone, PRL mediates hydromineral balance of euryhaline fish acclimated to freshwater (Pickford and Phillips, 1959), which is different from our results. As is well known, the PAT1 gene works in the absorption of Cl– in gills (Lee et al., 2006), which is consistent with transcriptome sequencing data and the results of real-time quantitative PCR. Apolipoprotein (APO) is the major protein component of high-density lipoprotein particles in serum and participates in the reverse transport of cholesterol from tissues to the liver for excretion (Fielding et al., 1972). Considering that fish exploit lipids as the major energy source (after proteins), lipid metabolism is important for osmoregulation and homeostasis in fish. The amplification of gene APOM mRNA was found to be highest in gill, liver, and kidney, moderate in spleen, and weak in other tissues. For gene HMDH, the expression levels in spleen and kidney were significantly higher than in other tissues, which proved the obvious tissue specificity of gene HMDH expression and the importance of kidney and spleen in osmotic pressure regulation. The functions of HMDH in vertebrate endocrine systems (Goward and Nicholls, 1994) is consistent with that of turbot transcriptome analyses, which showed that the gene encoding core proteins of renin secretion changed. In adult zebrafish, gene FABP6 was expressed in intestine, ovary, liver, heart, and kidney (Alves-Costa et al., 2008). Our results showed that the expression of gene FABP6 mRNA was higher in the gills, liver, spleen, and kidney, and was low in other tissues, presumably because of the fish species and tissue specificity of gene expression. After our analyses, we hypothesized that the kidney, gill, and spleen were vital regulating organs of osmotic pressure, and the species specificity of genes related with osmoregulation expression showed that the osmoregulation pattern of euryhaline fishes differed between species.
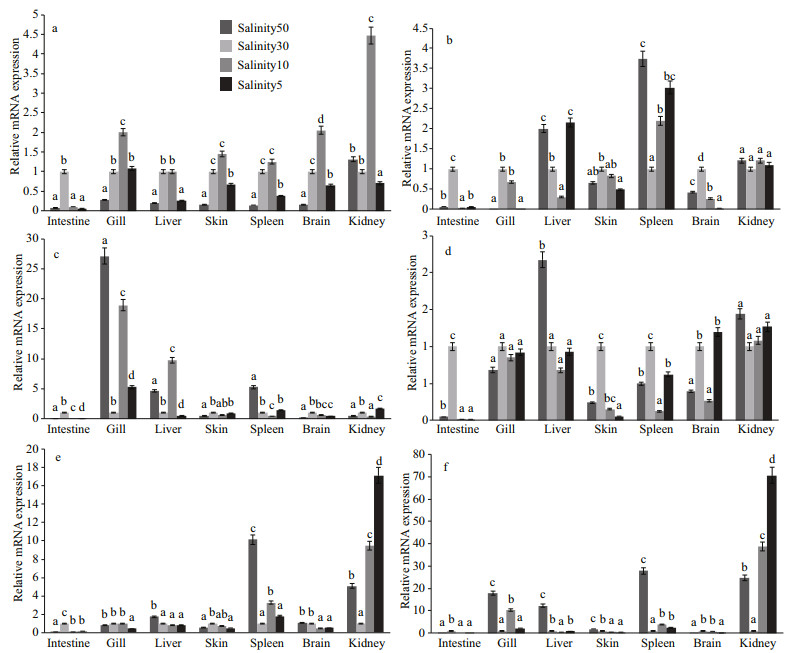
|
| Fig.9 Expression of target genes in seven tissues of S. maximus after osmotic stress Different letters represent significant differences (P < 0.05) between the columns. a. NKAα1 (Na+-K+-ATPaseα1); b.PRL (prolactin); c. PAT1 (solute carrier family 26 member 6, SLC26A6); d. APOM (apolipoprotein M); e. HMDH (3-hydroxyl-3-methylglutaryl-coenzyme A reductase); f. FABP6 (fatty acidbinding protein 6). |
The kidney transcriptome of turbot responded to brackish water was sequenced and characterized. In silico analysis indicated that the sequence of transcripts of various abundance and length were represented in the assembled contigs. The functional annotation analysis of transcriptomes showed that the immune system and the biological processes associated with digestion, absorption, and metabolism played an important role in osmoregulation in turbot responded to hypo-salinity. The biological processes of mineral absorption and bile secretion contributed to iono-osmoregulation and resulted in cell volume regulation and cell phenotypic plasticity. Of importance, we have analyzed and predicted signal transduction networks composed of the MAPK signaling pathway and PI3K-Akt signaling pathway with the GABAergic synapse worked in hypoosmoregulation signal transduction in turbot. To the best of our knowledge, this was the first NGS study that provided a transcriptome-wide perspective about the mechanisms of osmoregulation in kidneys of turbot responded to hypo-salinity. In addition, an analysis of the tissue specific expression profiles of targeted genes subjected to salinity stress showed that the kidney, gill, and spleen were vital as regulating organs under osmotic pressure, and the osmoregulation patterns of euryhaline fishes differed between species. Consistent with other studies, our studies indicated that many of the genes and signal pathways related with iono-osmoregulation were identified in the view of transcriptome. Finally, this study has provided an unprecedented global transcriptomic view of kidney iono-osmoregulation.
5 DATA AVAILABILITY STATEMENTThe raw reads for the next generation sequencing data have been deposited in NCBI Sequence Read Archive (SRA) with the GenBank number SRP153594.
Alves-Costa F A, Denovan-Wright E M, Thisse C, Thisse B, Wright J M. 2008. Spatio-temporal distribution of fatty acid-binding protein 6 (fabp6) gene transcripts in the developing and adult zebrafish (Danio rerio). FEBS J., 275(13): 3 325-3 334.
DOI:10.1111/j.1742-4658.2008.06480.x |
Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol., 11(10): R106.
DOI:10.1186/gb-2010-11-10-r106 |
Arden K C. 2004. FoxO:linking new signaling pathways. Mol.Cell, 14(4): 416-418.
DOI:10.1016/S1097-2765(04)00213-8 |
Babey M, Kopp P, Robertson G L. 2011. Familial forms of diabetes insipidus:clinical and molecular characteristics. Nat. Rev. Endocrinol., 7(12): 701-714.
DOI:10.1038/nrendo.2011.100 |
Bakir B, Sezerman O U. 2006. Functional classification of G-protein coupled receptors, based on their specific ligand coupling patterns. In: Rothlauf F, Branke J, Cagnoni S, Costa E, Cotta C, Drechsler R, Lutton E, Machado P, Moore J H, Romero J, Smith G D, Squillero G, Takagi H eds. Applications of Evolutionary Computing. Springer, Berlin, Heidelberg. p.1-12.
|
Beyenbach K W. 1986. Secretory NaCl and volume flow in renal tubules. Am. J. Physiol., 250(5): R753-R763.
|
Beyenbach K W. 2004. Kidneys sans glomeruli. Am. J. Physiol.Renal Physiol., 286(5): F811-F827.
DOI:10.1152/ajprenal.00351.2003 |
Bhattacharya S. 2004. Handbook of cell signaling. J. Anat., 205(1): 77.
DOI:10.1111/j.0021-8782.2004.00308.x |
Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank D A, Rozovsky I, Stahl N, Yancopoulos G D, Greenberg M E. 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science, 278(5337): 477-483.
DOI:10.1126/science.278.5337.477 |
Brennan R S, Galvez F, Whitehead A. 2015. Reciprocal osmotic challenges reveal mechanisms of divergence in phenotypic plasticity in the killifish Fundulus heteroclitus. J. Exp. Biol., 218(8): 1 212-1 222.
DOI:10.1242/jeb.110445 |
Cutler C P, Cramb G. 2002. Branchial expression of an aquaporin 3 (AQP-3) homologue is downregulated in the European eel Anguilla anguilla following seawater acclimation. J. Exp. Biol., 205(17): 2 643-2 651.
|
Cutler C P, Philips C, Hazon N, Cramb G. 2009. Aquaporin 8(AQP8) intestinal mRNA expression increases in response to salinity acclimation in yellow and silver European eels(Anguilla anguilla). Comp. Biochem. Physiol. A Mol.Integr. Physiol., 153(S2): S78.
DOI:10.1016/j.cbpa.2009.04.047 |
Dietz C, Stiller K T, Griese M, Schulz C, Susenbeth A. 2013. Influence of salinity on energy metabolism in juvenile turbot, Psetta maxima (L.). Aquacult. Nutr., 19(S1): 135-150.
DOI:10.1111/anu.12034 |
Edwards S L, Marshall W S. 2012. Principles and patterns of osmoregulation and euryhalinity in fishes. Fish Physiol., 32: 1-44.
DOI:10.1016/B978-0-12-396951-4.00001-3 |
Ehebauer M, Hayward P, Martinez-Arias A. 2006. Notch signaling pathway. Sci. STKE, 2006(364): m7.
DOI:10.1126/stke.3642006cm7 |
Farrell A P. 2011. Encyclopedia of Fish Physiology: from Genome to Environment. Academic Press, San Diego.
|
Fielding C J, Shore V G, Fielding P E. 1972. A protein cofactor of lecithin:cholesterol acyltransferase. Biochem. Biophys.Res. Commun., 46(4): 1 493-1 498.
DOI:10.1016/0006-291X(72)90776-0 |
Fiol D F, Kültz D. 2007. Osmotic stress sensing and signaling in fishes. FEBS J., 274(22): 5 790-5 798.
DOI:10.1111/j.1742-4658.2007.06099.x |
Fouchs A, Ollivier H, Haond C, Roy S, Calvès P, PichavantRafini K. 2010. Activation of the MAPKs ERK1/2 by cell swelling in turbot hepatocytes. Biol. Cell, 102(8): 447-456.
DOI:10.1042/BC20090154 |
Götz S, García-Gómez J M, Terol J, Williams T D, Nagaraj S H, Nueda M J, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the blast2go suite. Nucleic Acids Res., 36(10): 3 420-3 435.
DOI:10.1093/nar/gkn176 |
Goward C R, Nicholls D J. 1994. Malate dehydrogenase:a model for structure, evolution, and catalysis. Protein Sci., 3(10): 1 883-1 888.
DOI:10.1002/pro.5560031027 |
Grabherr M G, Haas B J, Yassour M, Levin J Z, Thompson D A, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q D, Chen Z H, Mauceli E, Hacohen N, Gnirke A, Rhind N, Di Palma F, Birren B W, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Trinity:reconstructing a fulllength transcriptome without a genome from RNA-Seq data. Nat. Biotechnol., 29(7): 644-652.
DOI:10.1038/nbt.1883 |
Grant S, Qiao L, Dent P. 2002. Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front Biosci., 7: d376-389.
|
Hoffmann E K, Lambert I H, Pedersen S F. 2009. Physiology of cell volume regulation in vertebrates. Physiol. Rev., 89(1): 193-277.
DOI:10.1152/physrev.00037.2007 |
Hummel C S, Lu C, Loo D D F, Hirayama B A, Voss A A, Wright E M. 2011. Glucose transport by human renal Na+/D-glucose cotransporters SGLT1 and SGLT2. Am. J.Physiol. Cell Physiol., 300(1): C14-C21.
DOI:10.1152/ajpcell.00388.2010 |
Imsland A K, Foss A, Gunnarsson S, Berntssen M H G, FitzGerald R, Bonga S W, v Ham E, Nævdal G, Stefansson S O. 2001. The interaction of temperature and salinity on growth and food conversion in juvenile turbot(Scophthalmus maximus). Aquaculture, 198(3-4): 353-367.
DOI:10.1016/S0044-8486(01)00507-5 |
Inokuchi M, Hiroi J, Watanabe S, Lee K M, Kaneko T. 2008. Gene expression and morphological localization of NHE3, NCC and NKCC1a in branchial mitochondria-rich cells of mozambique tilapia (Oreochromis mossambicus)acclimated to a wide range of salinities. Comp. Biochem.Physiol. A Mol. Integr. Physiol., 151(2): 151-158.
DOI:10.1016/j.cbpa.2008.06.012 |
Kalujnaia S, McWilliam I S, Zaguinaiko V A, Feilen A L, Nicholson J, Hazon N, Cutler C P, Cramb G. 2007. Transcriptomic approach to the study of osmoregulation in the European eel Anguilla anguilla. Physiol. Genomics, 31(3): 385-401.
DOI:10.1152/physiolgenomics.00059.2007 |
Karaica D, Breljak D, Brzica H, Lončar J, Herak-Kramberger C M, Micek V, Vrhovac I, Ivković D J, Mihaljević I, Marić P, Smital T, Burckhardt B C, Burckhardt G, Sabolić I. 2015. CFEX (slc26a6) in rat kidneys, liver, and small intestine in an experimental model of oxalate nephrolitiasis. Arch. Ind. Hyg. Toxicol., 66(3): 228.
|
Kasprowicz A. 2011. Osmosensing. In: Wojtaszek P ed.Mechanical Integration of Plant Cells and Plants. Springer, Berlin, Heidelberg. p.225-240.
|
Kültz D, Avila K. 2001. Mitogen-activated protein kinases are in vivo transducers of osmosensory signals in fish gill cells. Comp. Biochem. Physiol. B Biochem. Mol. Biol., 129(4): 821-829.
DOI:10.1016/S1096-4959(01)00395-5 |
Kültz D, Burg M. 1998. Evolution of osmotic stress signaling via MAP kinase cascades. J. Exp. Biol., 201(22): 3 015-3 021.
|
Kültz D. 2015. Physiological mechanisms used by fish to cope with salinity stress. J. Exp. Biol., 218(12): 1 907-1 914.
DOI:10.1242/jeb.118695 |
Kurt B, Kurtz L, Sequeira-Lopez M L, Gomez R A, Willecke K, Wagner C, Kurtz A. 2011. Reciprocal expression of connexin 40 and 45 during phenotypical changes in reninsecreting cells. Am. J. Physiol. Renal Physiol., 300(3): F743-F748.
DOI:10.1152/ajprenal.00647.2010 |
Kurtz A, Wagner C. 1999. Cellular control of renin secretion. J. Exp. Biol., 202(3): 219-225.
|
Lam S H, Lui E Y, Li Z J, Cai S J, Sung W K, Mathavan S, Lam T J, Ip Y K. 2014. Differential transcriptomic analyses revealed genes and signaling pathways involved in ionoosmoregulation and cellular remodeling in the gills of euryhaline mozambique tilapia, Oreochromis mossambicus. BMC Genomics, 15(1): 921.
DOI:10.1186/1471-2164-15-921 |
Lee K M, Kaneko T, Katoh F, Aida K. 2006. Prolactin gene expression and gill chloride cell activity in fugu Takifugu rubripes exposed to a hypoosmotic environment. Gen.Comp. Endocrinol., 149(3): 285-293.
DOI:10.1016/j.ygcen.2006.06.009 |
Li B, Dewey C N. 2011. Rsem:accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12: 323.
DOI:10.1186/1471-2105-12-323 |
Mao X Z, Cai T, Olyarchuk J G, Wei L P. 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 21(19): 3 787-3 793.
DOI:10.1093/bioinformatics/bti430 |
Marshall W, Grosell M. 2005. Ion transport, osmoregulation, and acid-base balance. In: Evans D H, Claiborne J B eds.The Physiology of Fishes. 3rd ed. CRC Press, Boca Raton.p.177-230.
|
Narnaware B Y K, Kelly S P, Woo N Y S. 2000. Effect of salinity and ration size on macrophage phagocytosis in juvenile black sea bream (Mylio macrocephalus). J. Appl.Ichthyol., 16(2): 86-88.
DOI:10.1046/j.1439-0426.2000.00113.x |
Nishimoto G, Sasaki G, Yaoita E, Nameta M, Li H, Furuse K, Fujinaka H, Yoshida Y, Mitsudome A, Yamamoto T. 2007. Molecular characterization of water-selective AQP (EbAQP4)in hagfish:insight into ancestral origin of AQP4. Am. J.Physiol. Regul. Integr. Comp. Physiol., 292(1): R644-R651.
DOI:10.1152/ajpregu.00362.2006 |
Oelkers P, Kirby L C, Heubi J E, Dawson P A. 1997. Primary bile acid malabsorption caused by mutations in the ileal sodium-dependent bile acid transporter gene (SLC10A2). J. Clin. Invest., 99(8): 1 880-1 887.
DOI:10.1172/JCI119355 |
Pereiro P, Balseiro P, Romero A, Dios S, Forn-Cuni G, Fuste B, Planas J V, Beltran S, Novoa B, Figueras A. 2012. Highthroughput sequence analysis of turbot (Scophthalmus maximus) transcriptome using 454-pyrosequencing for the discovery of antiviral immune genes. PLoS One, 7(5): e35369.
DOI:10.1371/journal.pone.0035369 |
Pickford G E, Phillips J G. 1959. Prolactin, a factor in promoting survival of hypophysectomized killifish in fresh water. Science, 130(3373): 454-455.
DOI:10.1126/science.130.3373.454 |
Prodocimo, V, Souza C F, Pessini C, Fernandes L C, Freire C A. 2008. Metabolic substrates are not mobilized from the osmoregulatory organs (gills and kidney) of the estuarine pufferfishes Sphoeroides greeleyi and S. testudineus upon short-term salinity reduction. Neotrop. Ichthyol., 6(4): 613-620.
DOI:10.1590/S1679-62252008000400009 |
Rappolee D A, Armant D R. 2009. Cell signaling. In: Krawetz S ed. Bioinformatics for Systems Biology. Humana Press, New York. p.89-104.
|
Schmittgen T D, Zakrajsek B A. 2000. Effect of experimental treatment on housekeeping gene expression:validation by real-time, quantitative RT-PCR. J. Biochem. Biophys.Methods, 46(1-2): 69-81.
DOI:10.1016/S0165-022X(00)00129-9 |
Shibata Y, Kumai M, Nishii K, Nakamura K. 2001. Diversity and molecular anatomy of gap junctions. Med. Electron Microsc., 34(3): 153-159.
DOI:10.1007/s007950100008 |
Tang X M, Sui Z, Tian J B, Wang G F. 2006. Effects of salinity on metabolic rate of juvenile turbot (Scophamus maximus). South China Fish. Sci., 2(4): 54-58.
(in Chinese with English abstract) |
Trapnell C, Williams B A, Pertea G, Mortazavi A, Kwan G, Van Baren M J, Salzberg S L, Wold B J, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol, 28(5): 511-515.
DOI:10.1038/nbt.1621 |
Vojtek A B, Der C J. 1998. Increasing complexity of the Ras signaling pathway. J. Biol. Chem., 273(32): 19.
|
Xu Z Z, Gan L, Li T Y, Xu C, Chen K, Wang X D, Qin J G, Chen L Q, Li E C. 2015. Transcriptome profiling and molecular pathway analysis of genes in association with salinity adaptation in Nile tilapia Oreochromis niloticus. PLoS One, 10(8): e0136506.
DOI:10.1371/journal.pone.0136506 |
Young M D, Wakefield M J, Smyth G K, Oshlack A. 2010. Gene ontology analysis for RNA-seq:accounting for selection bias. Genome Biol., 11(2): R14.
DOI:10.1186/gb-2010-11-2-r14 |
 2020, Vol. 38
2020, Vol. 38