Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CAO Liang, SONG Xuelin, ZHANG E
- The first mitogenome of the Nile pufferfish Tetraodon lineatus from Lake Turkana in East Africa: new insights into the genus
- Journal of Oceanology and Limnology, 38(2): 490-502
- http://dx.doi.org/10.1007/s00343-019-9050-8
Article History
- Received Mar. 6, 2019
- accepted in principle Apr. 28, 2019
- accepted for publication Aug. 13, 2020
2 University of Chinese Academy of Sciences, Beijing 100049, China;
3 Sino-Africa Joint Research Center, Chinese Academy of Sciences, Wuhan 430074, China
The family Tetraodontidae, also popularly known as pufferfish, is the most speciose group of the order Tetraodontiformes and is widespread across many tropical and subtropical regions of the world. It includes species that are typically marine, with occasional entering and occurring in brackish and freshwaters (Nelson, 2006). Pufferfishes are notable for their conspicuous defensive behavior: when threatened, they inflate the body to a volume nearly four times larger than normal size (Brainerd, 1994). These fishes are also remarkable for being the smallest genomes among vertebrates, approximately 400 MB or 1/8 the size of the human genome (Hinegardner and Rosen, 1972). Thus, two pufferfish species, namely Dichotomyctere nigroviridis and Takifugu rubripes, have been selected as model organisms to study the evolution of vertebrate genomes (Brenner et al., 1993; Crnogorac-Jurcevic et al., 1997).
The genus Tetraodon Linnaeus, 1758, as traditionally conceived, is a group of pufferfish widely occurring in tropical African and Asian regions where they are dwellers of marine and freshwater systems, particularly in estuaries and inshore waters (Kottelat, 2013). A total of about 24 pufferfish species have been designated to this genus. From Kottelat's (2013) point of view, Tetraodon s.s. is an endemic African genus including six valid species: T. duboisi, T. lineatus, T. mbu, T. miurus, T. pustulatus, and T. schoutede. Among them, T. lineatus is the most widespread, ranging from the Senegal and Volta River basins to the Niger and Nile River basins as far as to Lake Turkana, while T. pustulatus is confined to the Cross River basin in southeastern Nigeria. All other four species are so far known from the Congo River basin (Ebert, 2001).
Recently, the Nile pufferfish T. lineatus was reported from Shanghai City, East China, and its complete mitogenome was determined and its phylogenetic relationship with other two African pufferfishes was inferred based on mitogenome (Gong et al., 2016). Undoubtedly, this specimen found in a Chinese coastal location is completely outside T. lineatus' known distribution in Africa. The report on the occurrence of this African pufferfish in China is not free from the question. The clarification of the source and species recognition of Chinese sample is thus warranted. Molecular technology can help to find solutions to these issues when fresh samples of T. lineatus from the African continent are available.
The non-monophyletic nature of Tetraodon s.l. was unveiled in many molecular phylogenetic analyses of this genus and the family Tetraodontidae (Yamanoue et al., 2011; Igarashi et al., 2013; Santini et al., 2013). In these analyses, all species of Tetraodon clustered into three distantly allied groups: Asian freshwater group, Asian brackish water group, and African freshwater group. This is one of the factors that led Kottelat (2013) to restrict Tetraodon s.s. to African freshwater group. Nonetheless, molecular evidence for the monophyletic nature of the genus is still not totally conclusive. The crucial reason for this is that T. lineatus, the type species of the genus, was either not included or included but ambiguously identified in previous molecular studies.
Lake Turkana, the world's largest permanent alkaline desert lake located in eastern Rift Valley in East Africa, is an endorheic lake situated in northern Kenya with its far northern end crossing into southern Ethiopia. It is home to around 50 fish species (Nyingi, 2013), most of which are present in the Nile River basin (Wakjira and Getahun, 2017). There is a common consensus that the Nilotic-Sudanic fish fauna historically extended into the eastern Rift Valley lakes inclusive of Lake Turkana (Roberts, 1975). This lake, however, has its own unique fish species, with a total of 11 endemics (Hardman, 2008; Nyingi, 2013).
Lake Turkana falls within the known distribution of T. lineatus, and this species is the only pufferfish presently reported from this Lake. During a fish survey conducted by us in August, 2016 in Lake Turkana, Kenya, a single pufferfish specimen was collected. We amplified its complete mitochondrial genome and inferred its phylogenetic relationship with other pufferfishes. The aims of the present study are to confirm the morphology-based recognition of the pufferfish sample from Lake Turkana as T. lineatus utilizing molecular evidence, verify the source and species identification of the Nile pufferfish sample recently reported from China and address issues regarding the monophyletic nature of Tetraodon s.s.
2 MATERIAL AND METHOD 2.1 Taxon sampling and DNA extractionOne specimen of the Nile pufferfish from Lake Turkana in Kenya (Fig. 1) was collected in August, 2016. Diagnostic characters provided by Lévêque (1992) for T. lineatus includes: no scales present on body, but head and body covered with small spines except on snout and caudal peduncle; lateral line absent, a pair of fused teeth at front of each jaw; two pairs of non-perforated nasal tentacles; nostrils consisting of two fleshy lobes located in front of folded collar surrounding the opening; dorsal and anal fins short, placed far back on body; pectoral fins well-developed; no pelvic fins; caudal fin truncate to rounded; yellow longitudinal stripes along sides of body in adults, but black-rimmed red ocelli (eyespots) present in juveniles. All these characters are shared with the pufferfish specimen collected from Lake Turkana in Kenya, so the specimen was identified as T. lineatus. See Fig. 1 for the picture of this species captured by us from Lake Turkana; its coloration is similar to Nyingi's (2013) pictures.

|
| Fig.1 Lateral view of the Nile pufferfish Tetraodon lineatus, IHB 2016077390, 199.5 mm SL, collected from Lake Turkana in Kenya |
The tissue (right pectoral-fin tip) employed for DNA extraction was sampled from the fresh specimen immediately after capture and stored in 95% ethanol. Genomic DNA was extracted from a small piece of the pectoral-fin clip (30 mg) using a TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing), following the manufacturer's instructions. The voucher specimen (IHB 2016077390) is kept in the Freshwater Organism Museum at the Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei Province, China.
2.2 PCR and sequencingThe complete mitogenome was amplified in three overlapping segments by the long PCR technique (Lo, 1998). Software Primer Premier 5.0 was used for primer design (Singh et al., 1998; Lalitha, 2000). Long-range PCR primers were designed based on three versatile primers, which were used to amplify 16SrRNA (16Sar-L/16Sbr-H), COI (FishF1/FishR1) and Cyt b (Cytball-Fl/ Cytball-Rl) segments. These three short sequences were utilized as templates to design three long-primers: Lincy16H/Lincy16L (amplification product was the fragment from Cyt b to 16SrRNA), Lin16COH/Lin16COL (amplification product was the fragment from 16SrRNA to COI), and LinCOCYH/LinCOCYL (amplification product was the fragment from COI to Cyt b) (Table 1). For the three short sequences, the PCR was performed in a total reaction volume of 25 μL containing 2.5 μL of 10×PCR buffer, 1.5 μL dNTPs (2.5 mmol/L), 0.5 μL of each primer (10 mmol/L), 0.5 μL rTaq (5 U/μL) and 1 μL genomic DNA template, and sterilized water was added to the PCR mixture to bring the final volume to 25 μL. For the three long-range segments, the PCR were conducted in 50 μL volume including 5 μL 10×PCR buffer, 8 μL dNTPs (2.5 mmol/L), 0.5 μL of each primer (10 mmol/LM), 0.5 μL LA Taq (5 U/μL), 1 μL genomic DNA template, and sterilized water was added to the PCR mixture to bring the final volume to 50 μL. The thermocycling conditions for these primers were as follows: initial denaturation for 5 min at 94℃, denaturation for 50 s at 94℃, annealing for 50 s appropriate temperatures (Table 1), extension for 1 min/kb amplification according to the product length at 72℃. After 35 cycles, final extension was done at 72℃ for 10 min, and then the product was stored at 4℃. The PCR fragments were sequenced using the primer-walking strategy.

|
DNA sequences were assembled by SeqMan program (DNAstar), checked, and edited manually. Annotation of 13 protein-coding genes was completed by comparing with closely associated species within Tetraodon s.l. in ORF finder tool of NCBI (https://www.ncbi.nlm. nih. gov/orffinder/). The position and secondary structures of 22 tRNAs were determined by tRNAscan 2.0 (http://lowelab.ucsc.edu/tRNAscanSE/). For the small and large subunit ribosomal RNA genes (12SrRNA and 16SrRNA), the start and stop base pair were assumed adjacent to the ends of their neighboring genes. The same method was utilized for two non-coding regions, D-loop and the replication origin of L strand (OL). The former was defined as the sequence bounded by the genes for tRNA-Pro and tRNA-Phe, and the latter is a small fragment more than 30 bp in length and located between tRNA-Asn and tRNA-Cys (Bibb et al., 1981; Zhou et al., 1993).
2.4 Phylogenetic analysisFour datasets were used for phylogenetic analysis: the complete mitogenomes, COI, Cyt b and 16SrRNA sequences (Table 2). For the complete mitogenome dataset, there were 52 mitogenomes of Tetraodontiformes available in GenBank (Table 2). Three partial segments of COI, Cyt b and 16SrRNA gene were separated from 52 available complete mitogenomes in order to derive phylogenetic relationship based on these genes. Single gene's sequences of the three partial segments available in GenBank were added, reference to Table 2. The generic classification of the family Tetraodontidae given by Kottelat (2013) was followed.
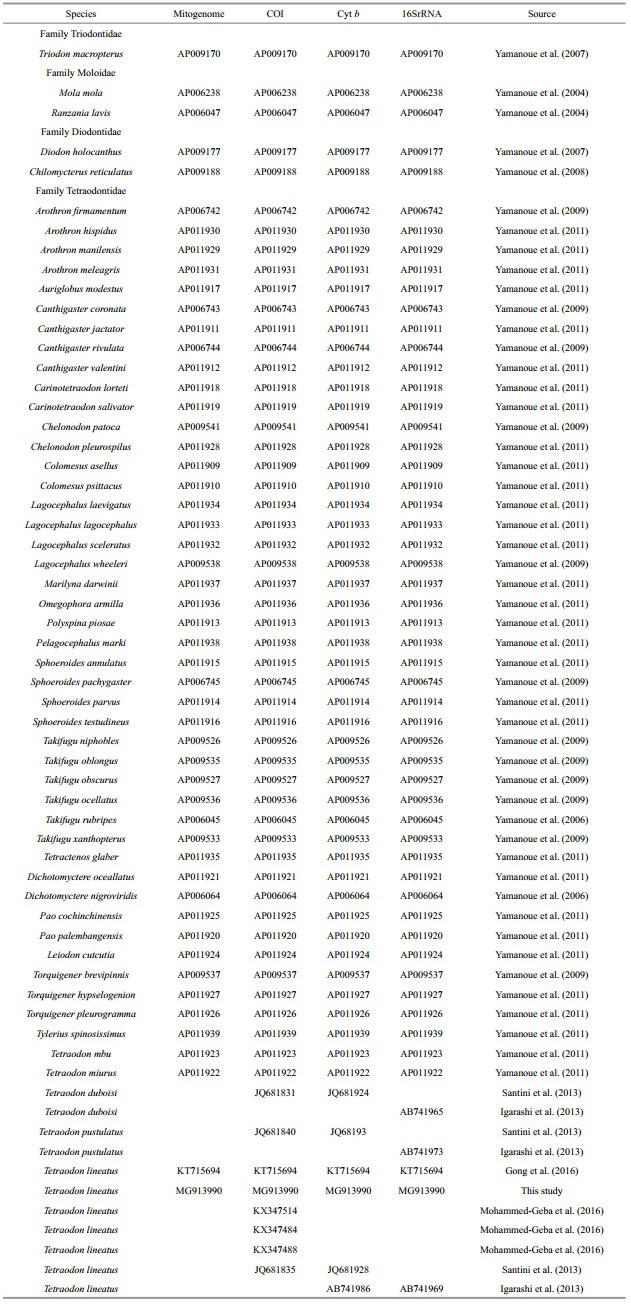
|
DNA sequences were translated into protein sequences to check if nonsense mutation occurred (Li et al., 2008). Sequences coding start- and stop-codons were excluded to eliminate homogeneity. Transition and transversion on the three codon positions were plotted against F84 genetic distance by DAMBE (Xia and Xie, 2001) to assess the saturation of the 13 genes. Sequences were aligned using MAFFT v7.407 (Katoh et al., 2002) and then aggregated into a sequence matrix. Phylogenetic trees were inferred by raxmlGUI for maximum-likelihood tree (Silvestro and Michalak, 2012) by the ML+rapid bootstrap searching method and GTRGAMMA model for 1000 repetitions. Evolutionary models were chosen through PartitionFinder V1.1.1 (Lanfear et al., 2012) on the basis of AIC criterion, setting partition by codon for each gene and gene for the combined dataset. Final rooting was done utilizing Triodon macropterus (Triodontidae), Mola mola (Molidae) and Diodon holocanthus (Diodontidae), based on Yamanoue et al.'s (2011) results.
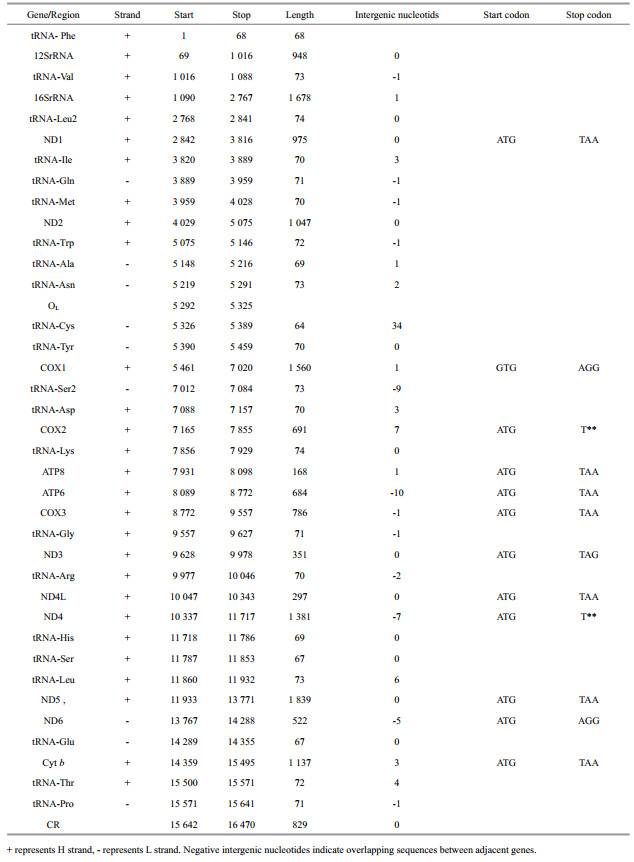
3 RESULT 3.1 Genome organizationThe complete mitochondrial genome of T. lineatus from Lake Turkana was 16 470 bp in length (GenBank No. MG913990) (Fig. 2, Table 3). The mitogenome of this species, just like other four pufferfishes of the genus Tetraodon s.s., consisted of 37 genes (for genes codes, please refer to legend in Fig. 2), 13 of which are protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes and two non-coding regions, as found in other vertebrate animals (Wang et al., 2011a, b). Out of these, 28 encoded on the H-strand, and the remaining nine genes encoded on the L-strand, including one protein coding gene (ND6) and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro).
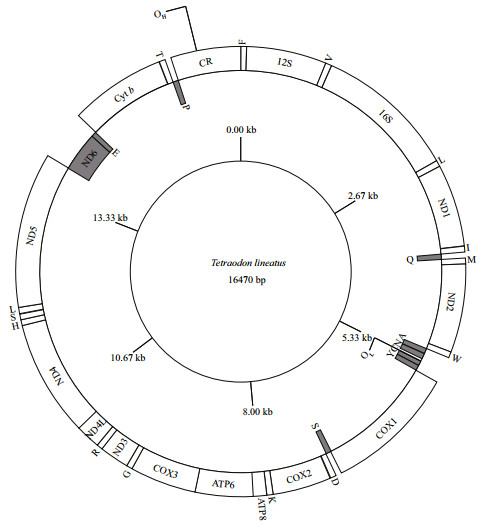
|
| Fig.2 Mitogenomic organization of the Nile pufferfish Tetraodon lineatus from Lake Turkana in Kenya he L-strand encoding genes are shown in shaded boxes and labeled inside of the circle. Genes are abbreviated as follows: ATP6 and ATP8 (subunits 6 and 8 of ATPase), COX1-3 (cytochrome oxidase subunits 1-3), Cyt b (cytochrome b), ND1-6 and ND4L (NADH dehydrogenase subunits 1–6 and 4L), 12S and 16S (ribosomal RNA of 12S and 16S), CR (control region). One-letter amino acid abbreviations were used to label the corresponding tRNA genes. OH and OL indicate the origins of H- and L- strand replication. |
Ten intergenic spacers were recognized, ranging from 1–6 bp in size. Gene overlaps varying from 1–10 bp were observed in 13 regions, including four notable overlapping positions between genes: ATP6 and ATP8, ND4 and ND4L, ND5 and ND6, COI and tRNA-Ser. The first three overlaps have been also reported from other fish species (Yu and Kwak, 2015). ATG is the most frequently used start codon for the 13 protein-coding genes, with a unique exception, GTG utilized by COI gene. GTG start codon for the COI gene is also present in other fish species (Wang et al., 2011a, b; Yu and Kwak, 2015). Eight genes' (ND1, ND2, ATP8, ATP6, COIII, ND4L, ND5, Cyt b) stop codon is TAA, while two genes' (COI and ND6) stop codons are AGG, and ND3's is TAG. Both ND4 and COII exhibit incomplete stop codons with a single "T" residue. The presence of incomplete stop codons is common in fish species mitochondrial genes (Peng et al., 2006; Kartavtsev et al., 2007; Yu and Kwak, 2015).
The complete mitochondrial genome of T. lineatus from Lake Turkana was 11 bp longer than the previously published one for a sample (KT715694, 16 459 bp in length) recognized as T. lineatus from China by Gong et al. (2016). The overall nucleotide sequence similarity between these two mitogenomes was 98%. The 13 protein-coding genes and two rRNA genes exhibited 95%–100% similarities in nucleotide sequence; major length distinction came from 16SrRNA gene and the control region.
3.2 Sequence variationsThe pairwise transition and transversion differences for each codon increased with F84 genetic distance, except TS difference of the third codon which asymptotically approached to saturation at about 0.4 distance and increases were hardly observed (Fig. 3). Nucleotide frequencies at all codon positions differed between the two strands. On the H-strand, the nucleotide frequencies were C>A>G>T at the first codon position, T>C>A>G at the second codon position and C>A>T>G at the third codon position. Meanwhile, on the L-strand, the nucleotide frequencies were G>T>A>C at the first codon position, A>G>T>C at the second codon position and G>T>C>A at the third codon position.
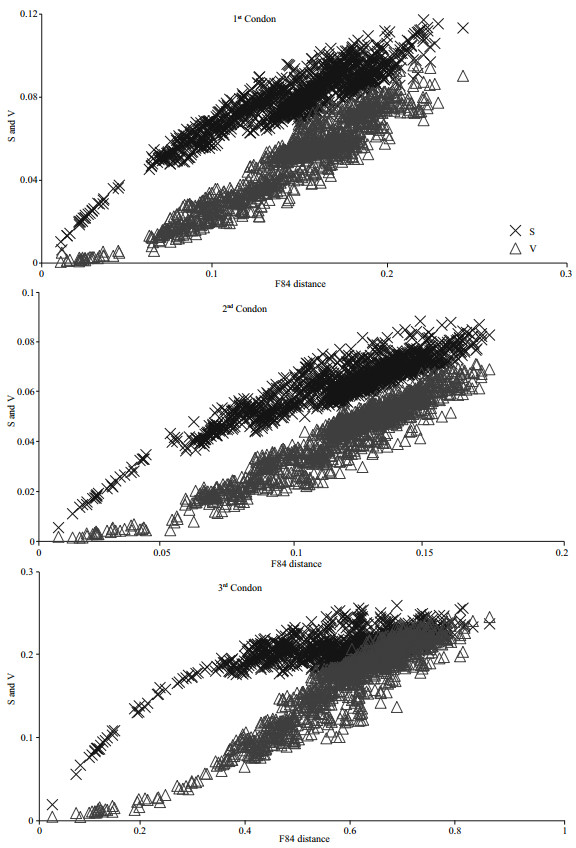
|
| Fig.3 Plot for transition and transversion on the three codon positions of the concatenated 13 protein coding genes (PCGs) versus pairwise F84 genetic distance of the 47 Tetraodontidae species and three outgroups S: transition; V: transversion. |
Phylogenetic trees generated from the complete mitogenome, COI, Cyt b, and 16SrRNA gene for the family Tetraodontidae were shown in Fig. 4. Since Tetraodon s.s. was the target group of this study, each sample of this genus used in this study was retained as the terminal taxon in these resultant phylogenetic trees where they were marked as red lines.
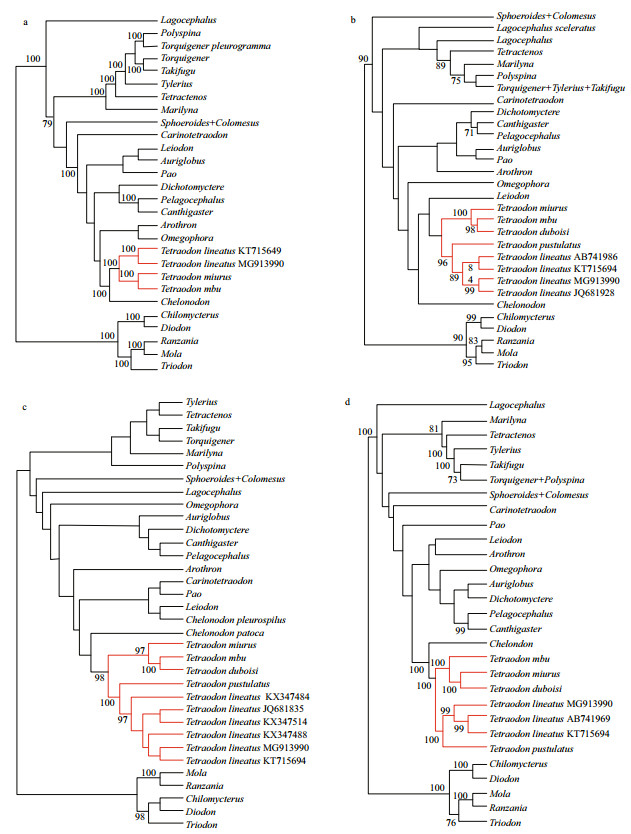
|
| Fig.4 The maximum-likelihood (ML) trees of Tetraodontidae inferred from a. the complete mitogenome; b. Cyt b; c. COI; d. 16SrRNA. Bootstrap values of ML (>70%) were shown at the nodes. Red lines indicate species belonging to the genus Tetraodon. |
In all trees constructed in this study, all analyzed samples of Tetraodon grouped together into a wellsupported monophyletic lineage (Fig. 4). The sample of the Nile pufferfish from Lake Turkana (MG913990) formed a well-supported monophyletic lineage, which represents T. lineatus (see the discussion below), with the sample (KT715694) from Shanghai, China in trees inferred from the complete mitogenome (Fig. 4a). In COI, Cyt b and 16SrRNA gene-based trees (Fig. 4b–d), samples of T. lineatus formed a monophyletic clade, which then clustered with T. pustulatus. These samples together formed a wellsupported clade with other three species of Tetraodon (T. mbu, T. miurus, and T. duboisi).
4 DISCUSSIONLake Turkana falls within the distribution of the Nile pufferfish T. lineatus based on known literatures. Deraniyagala (1948) was the first to describe the pufferfish of the lake as Tetraodon fahaka rudolfianus. The name was subsequently renamed to Tetraodon fahaka by Hamblyn (1962), Mann (1964), and Hopson and Hopson (1982). Seegers et al. (2003) synonymized T. fahaka with T. lineatus, a species initially described from the Nile River basin. This taxonomic treatment has thereafter been followed consistently by other authors (Habteselassie, 2012; Wakjira and Getahun, 2017). The pufferfish specimen collected from Lake Turkana in the Kenyan section shared with all the morphologic characters provided by Lévêque (1992). The coloration of this specimen is the same as Nyingi's (2013) and Wakjira and Getahun's (2017) pictures. Therefore, this pufferfish was identified as T. lineatus.
The complete mitogenome of the Nile pufferfish T. lineatus from Lake Turkana in Kenya was determined for the first time. This complete genome sequence has strongly supported the morphologybased recognition of the pufferfish specimen from this lake as T. lineatus. Its type locality is in the Nile River basin. Mohammed-Geba et al. (2016) analyzed COI gene genetic diversity of this species from the upper Nile River basin in Egypt. In our phylogenetic analysis based on a mitochondrial perspective, the Lake Turkana pufferfish sample clustered with those of the upper Nile River basin and one sample (JQ68 1835) without precise collecting location into a strongly-supported monophyletic lineage (Fig. 4c). Such a finding further supports the hypothesis that the specimen from Lake Turkana, whose mitochondrial genome was sequenced in this study, belongs to the Nile pufferfish species T. lineatus. There were three COI haplotypes of T. lineatus found by Mohammed-Geba et al. (2016) in 45 samples from three localities of the upper Nile River basin in Egypt. The most widespread haplotype among them was also shared with the pufferfish sample collected from Lake Turkana. The shared presence of the common haplotype in Lake Turkana and the upper Nile River basin provides molecular evidence in favor of the existing geological hypothesis of existed historical connections of both water systems (Feibel, 2011).
The mitogenome of T. lineatus from Lake Turkana from this study can be a reference genome to evaluate the recognition of the pufferfish specimen of this species by Gong et al. (2016) collected in Shanghai City, East China. In their phylogenetic trees inferred from the complete mitogenomes for the family Tetraodontidae, the Chinese pufferfish specimen was grouped with two African pufferfish species of Tetraodon to constitute a monophyletic lineage. Nevertheless, the identification of this sample as T. lineatus needs to be confirmed based on comparisons with African sample of this species, particularly from its type locality (in the Nile River basin). Our comparison found that there was a high similarity (98%) in complete mitogenomes between Gong et al.'s (2016) pufferfish sample and the T. lineatus specimen from Lake Turkana in Kenya. In complete mitogenome-based trees (Fig. 4a), these two samples were grouped with each other to form a lineage with four African species of Tetraodon. Based on a reanalysis of Yamanoue et al. (2011) dataset of pufferfishes, the genetic distance in complete mitogenome between both was 1.7%, less than the minimum one (2.6%) found between Sphoeroides annulatus and S. testudineus. These findings affirm that they are conspecific and belong to the same species T. lineatus.
Gong et al. (2016) reported that the Chinese specimen of T. lineatus was caught from Luchaogang, Shanghai, East China, without detailed information about its possible source. The Nile pufferfish has not previously been recorded in fish species list of China (Zhang et al., 2016) or Southeast Asia (Kottelat, 2013). However, Liang et al. (2006) listed T. lineatus as introduced fish species in Taiwan, China, pointing out that pet trade has served as the source of many exotic fish. Accordingly, Xiong et al. (2015) included T. lineatus on the species list of non-native freshwater fish in China. Our COI gene sequence comparison unveiled that the most common haplotype found by Mohammed-Geba et al. (2016) for samples of this species from the upper Nile River basin was also shared with Gong et al.'s (2016) sample. It is highly likely that their sample from Shanghai, China was of African origin. The introduction of the Nile pufferfish as pet fish from Africa is, therefore, a reasonable explanation for its presence in China.
The genus name Tetraodon was initially erected by Linnaeus in 1758, with T. lineatus as the type species. It has been widely considered as valid since the work by Tyler (1980). In previous molecular phylogenetic analyses of the family Tetraodontidae or the genus Tetraodon s.l. (Yamanoue et al., 2011; Igarashi et al., 2013; Santini et al., 2013), species of Tetraodon clustered into three distantly associated groups, thus indicating the non-monophyletic nature of the genus. Kottelat (2013) presented a re-classification for Tetraodon as traditionally diagnosed. His taxonomic classification was actually based on Yamanoue et al. (2011) and Igarashi et al. (2013) molecular trees, Dekkers' (1975) morphological data and Tyler's (1980) morphological descriptions, color pattern and its ontogeny (pers. comm., Kottelat, October, 2017). He restricted Tetraodon s.s. to the African freshwater group composed of six species: T. duboisi, T. lineatus, T. mbu, T. miurus, T. pustulatus, and T. schoutede. Nevertheless, molecular evidence for the monophyletic nature of this genus was strong but not totally conclusive. Only two species of African pufferfish (T. mbu and T. miurus) formed a lineage in Yamanoue et al.'s (2011) phylogenetic trees inferred from complete mitogenome. Unfortunately, the type species of the genus, T. lineatus, was not sampled. Five African pufferfish species were clustered together in Santini et al.'s (2013) phylogenetic analysis based on COI, Cyt b and six nuclear genes for the family Tetraodontidae. A monophyletic lineage formed by these five species was also recovered in Igarashi et al.'s (2013) molecular phylogenetic trees of the genus Tetraodon s.l. based on 16S rRNA and Cyt b gene. However, no information about the precise collecting locality of the voucher specimen of T. lineatus was provided. This species has a much wider distribution from the Senegal and Volta River basins to the Niger and Nile River basins and Lake Turkana. From taxonomists' point of view, the identification of a given species is doubtful if comparisons with its type or topotypic specimens are not made. In particular, COI gene comparisons found that the pufferfish sample from Lake Turkana was identical to those of the Nile River basin, the type locality of T. lineatus. In phylogenetic trees inferred from COI, Cyt b and 16SrRNA gene (Fig. 4b–d), the sample of Lake Turkana clustered with the samples of the species under the name of T. lineatus determined by Yamanoue et al. (2011), Santini et al. (2013) and Igarashi et al. (2013), into a monophyletic lineage which truly represents the species. The mitogenome amplified in this study for the pufferfish sample from Lake Turkana here recognized with confidence as T. lineatus can be employed as reference genome to clear up ambiguity in identification of this species in previous molecular studies and thus provide conclusive molecular evidence for the monophyletic nature of Tetraodon s.s.
5 CONCLUSIONThe complete mitogenome of the pufferfish specimen from Lake Turkana in Kenya supported the morphology-based recognition of the specimen as T. lineatus. Mitogenomic comparison and phylogenetic analysis provides conclusive molecular evidence for the monophyletic nature of Tetraodon s.s. and also for the identification of the Chinese specimen recently recorded as T. lineatus and its African source.
6 DATA AVAILABILITY STATEMENTThe complete mitochondrial genome presented here is deposited in GenBank (accession No. MG913990).
7 ACKNOWLEDGMENTWe would like to express thanks to the National Museums of Kenya (NMK) and the Kenya Wildlife Service (KWS) for facilitated surveys and issuance of research permits. Our thanks are also given to Joseph Gathua, Julius Kioko, and Brian Okwiri of Ichthyology Section, NMK for assistance with field work.
Bibb M J, Van Etten R A, Wright C T, Walberg M W, Clayton D A. 1981. Sequence and gene organization of mouse mitochondrial DNA. Cell, 26(2): 167-180.
DOI:10.1016/0092-8674(81)90300-7 |
Brainerd E L. 1994. Pufferfish inflation:functional morphology of postcranial structures in Diodon holocanthus(Tetraodontiformes). Journal of Morphology, 220(3): 243-261.
DOI:10.1002/jmor.1052200304 |
Brenner S, Elgar G, Sanford R, Macrae A, Venkatesh B, Aparicio S. 1993. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature, 366(6452): 265-268.
DOI:10.1038/366265a0 |
Crnogorac-Jurcevic T, Brown J R, Lehrach H, Schalkwyk L C. 1997. Tetraodon fluviatilis, a new puffer fish model for genome studies. Genomics, 41(2): 177-184.
DOI:10.1006/geno.1997.4646 |
Dekkers W J. 1975. Review of the Asiatic freshwater puffers of the genus Tetraodon Linnaeus, 1758 (Pisces, Tetraodontiformes, Tetraodontidae). Bijdragen tot de Dierkunde, 45(1): 87-142.
DOI:10.1163/26660644-04501006 |
Deraniyagala P E P. 1948. Some scientific results of two visits to Africa Spolia. Zeylanica, 25: 1-42.
|
Ebert K. 2001. The Puffers of Fresh and Brackish Waters.Aqualog verlag GmbH, Rodgau, Germany. 96p.
|
Feibel C S. 2011. A geological history of the Turkana basin. Evolutionary Anthropology:Issues, News, and Reviews, 20(6): 206-216.
DOI:10.1002/evan.20331 |
Gong X L, Zhang Q, Bao B L. 2016. Complete mitochondrial DNA sequence of globe fish Tetraodon lineatus (Linnaeus, 1758) and the phylogenetic analysis of Tetraodontidae. Mitochondrial DNA Part B, 1(2): 781-782.
|
Habteselassie R D. 2012. Fishes of Ethiopia: annotated checklist with pictorial identification guide. Ethiopian Fisheries and Aquatic Science Association, Addis Ababa, Ethiopia. 250p.
|
Hamblyn E. 1962. A note on Lake Rudolf East. African Freshwater Fisheries Research Organization Annual Report, 1961. p.46-47.
|
Hardman M. 2008. A new species of Chrysichthys (Siluriformes:Claroteidae) from Lake Turkana, Kenya. Proceedings of the Academy of Natural Sciences of Philadelphia, 157(1): 25-36.
DOI:10.1635/0097-3157(2008)157[25:ANSOCS]2.0.CO;2 |
Hinegardner R, Rosen D E. 1972. Cellular DNA content and the evolution of teleostean fishes. The American Naturalist, 106(951): 621-644.
DOI:10.1086/282801 |
Hopson A J, Hopson J. 1982. The fishes of Lake Turkana with a description of three new species: Alestes ferox sp. nov., Alestes minutus sp. nov. (Pisces, Characidae) and Barbus turkanae sp. nov. (Pisces, Cyprinidae). In: Hopson A J ed.Lake Turkana. Overseas Development Administration, London, England. p.283-347.
|
Igarashi Y, Doi H, Yamanoue Y, Kinoshita S, Ishibashi T, Ushio H, Asakawa S, Nishida M, Watabe S. 2013. Molecular phylogenetic relationship of Tetraodon pufferfish based on mitochondrial DNA analysis. Fisheries Science, 79(2): 243-250.
DOI:10.1007/s12562-013-0598-5 |
Kartavtsev Y P, Jung S O, Lee Y M, Byeon H K, Lee J S. 2007. Complete mitochondrial genome of the bullhead torrent catfish, Liobagrus obesus (Siluriformes, Amblycipididae):Genome description and phylogenetic considerations inferred from the Cyt b and 16S rRNA genes. Gene, 396(1): 13-27.
DOI:10.1016/j.gene.2007.01.027 |
Katoh K, Misawa K, Kuma K I, Miyata T. 2002. MAFFT:a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30(14): 3059-3066.
DOI:10.1093/nar/gkf436 |
Kottelat M. 2013. The fishes of the inland waters of southeast Asia:a catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. The Raffles Bulletin of Zoology, (S27): 1-663.
|
Lalitha S. 2000. Primer premier 5. Biotech Software & Internet Report, 1(6): 270-272.
|
Lanfear R, Calcott B, Ho S Y W, Guindon S. 2012. PartitionFinder:combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29(6): 1695-1701.
DOI:10.1093/molbev/mss020 |
Lévêque C. 1992. Tetraodontidae. In: Lévêque C, Paugy D, Teugels G G eds. The Fresh and Brackish Water Fishes of West Africa: Volume 2. Collection Faune Tropicale 28.MRAC, Tervuren, Belgique and ORSTOM, Paris, France.p.868-870.
|
Li Z Q, Guo B C, Li J B, He S P, Chen Y Y. 2008. Bayesian mixed models and divergence time estimation of Chinese cavefishes (Cyprinidae:Sinocyclocheilus). Chinese Science Bulletin, 53(15): 2342-2352.
|
Liang S H, Chuang L C, Chang M H. 2006. The pet trade as a source of invasive fish in Taiwan. Taiwania, 51(2): 93-98.
|
Lo Y M. 1998. Clinical Applications of PCR. Humana Press, Totowa, New Jersey, USA. 326p.
|
Mann M 1964. Report on A Fisheries Survey of Lake Rudolf, Kenya. Government printer, Nairobi, Kenya. Annual Report 1962/63: 53-62, East African Freshwater Fisheries Research Organization.
|
Mohammed-Geba K, Mahdy A A, Eissa A S A, Osman A G M. 2016. Analysis of population genetics of the endangered Nile pufferfish Tetraodon lineatus (Linnaeus, 1758) in the upper Egyptian River Nile. International Journal of Ecotoxicology and Ecobiology, 1(2): 60-66.
|
Nelson J S. 2006. Fishes of the world. John Wiley and Sons, New Jersey, USA. 601p.
|
Nyingi W D. 2013. Guide to Common Freshwater Fishes of Kenya. Moran Publishers, Nairobi, Kenya. 136p.
|
Peng Z G, Wang J, He S P. 2006. The complete mitochondrial genome of the helmet catfish Cranoglanis bouderius (Siluriformes:Cranoglanididae) and the phylogeny of otophysan fishes. Gene, 376(2): 290-297.
DOI:10.1016/j.gene.2006.04.014 |
Roberts T R. 1975. Geographical distribution of African freshwater fishes. Zoological Journal of the Linnean Society, 57(4): 249-319.
DOI:10.1111/j.1096-3642.1975.tb01893.x |
Santini F, Nguyen M T T, Sorenson L, Waltzek T B, Alfaro J W L, Eastman J M, Alfaro M E. 2013. Do habitat shifts drive diversification in teleost fishes? An example from the pufferfishes (Tetraodontidae). Journal of Evolutionary Biology, 26(5): 1003-1018.
DOI:10.1111/jeb.12112 |
Seegers L, De Vos L, Okeyo D O. 2003. Annotated checklist of the freshwater fishes of Kenya (excluding the lacustrine haplochromines from Lake Victoria). Journal of East African Natural History, 92(1): 11-47.
DOI:10.2982/0012-8317(2003)92[11:ACOTFF]2.0.CO;2 |
Silvestro D, Michalak I. 2012. raxmlGUI:a graphical frontend for RAxML. Organisms Diversity & Evolution, 12(4): 335-337.
|
Singh V K, Mangalam A K, Dwivedi S, Naik S. 1998. Primer premier:program for design of degenerate primers from a protein sequence. Biotechniques, 24(2): 318-319.
DOI:10.2144/98242pf02 |
Tyler J C. 1980. Osteology, Phylogeny, and Higher Classification of the Fishes of the Order Plectognathi(Tetraodontiformes). NOAA Technical Report NMFS Circular 434, NOAA, Washington DC, America. p.1-422.
|
Wakjira M, Getahun A. 2017. Ichthyofaunal diversity of the Omo-Turkana basin, East Africa, with specific reference to fish diversity within the limits of Ethiopian waters. Check List, 13(2): 2059.
DOI:10.15560/13.2.2059 |
Wang J J, Li P, Zhang Y G, Peng Z G. 2011a. The complete mitochondrial genome of Chinese rare minnow, Gobiocypris rarus (Teleostei:Cypriniformes). Mitochondrial DNA, 22(5-6): 178-180.
DOI:10.3109/19401736.2011.636441 |
Wang Y, Guo R, Li H, Zhang X Y, Du J, Song Z B. 2011b. The complete mitochondrial genome of the Sichuan taimen (Hucho bleekeri):repetitive sequences in the control region and phylogenetic implications for Salmonidae. Marine Genomics, 4(3): 221-228.
DOI:10.1016/j.margen.2011.06.003 |
Xia X, Xie Z. 2001. DAMBE:software package for data analysis in molecular biology and evolution. Journal of Heredity, 92(4): 371-373.
DOI:10.1093/jhered/92.4.371 |
Xiong W, Sui X Y, Liang S H, Chen Y F. 2015. Non-native freshwater fish species in China. Reviews in Fish Biology and Fisheries, 25(4): 651-687.
DOI:10.1007/s11160-015-9396-8 |
Yamanoue Y, Miya M, Doi H, Mabuchi K, Sakai H, Nishida M. 2011. Multiple invasions into freshwater by pufferfishes (Teleostei:Tetraodontidae):a mitogenomic perspective. PLoS One, 6(2): e17410.
DOI:10.1371/journal.pone.0017410 |
Yamanoue Y, Miya M, Inoue J G, Matsuura K, Nishida M. 2006. The mitochondrial genome of spotted green pufferfish Tetraodon nigroviridis (Teleostei:Tetraodontiformes) and divergence time estimation among model organisms in fishes. Genes & Genetic Systems, 81(1): 29-39.
|
Yamanoue Y, Miya M, Matsuura K, Katoh M, Sakai H, Nishida M. 2004. Mitochondrial genomes and phylogeny of the ocean sunfishes (Tetraodontiformes:Molidae). Ichthyological Research, 51(3): 269-273.
DOI:10.1007/s10228-004-0218-6 |
Yamanoue Y, Miya M, Matsuura K, Katoh M, Sakai H, Nishida M. 2008. A new perspective on phylogeny and evolution of tetraodontiform fishes (Pisces: Acanthopterygii) based on whole mitochondrial genome sequences: Basal ecological diversification? BMC Evolutionary Biology, 8: 212.
|
Yamanoue Y, Miya M, Matsuura K, Miyazawa S, Tsukamoto N, Doi H, Takahashi H, Mabuchi K, Nishida M, Sakai H. 2009. Explosive speciation of Takifugu:another use of fugu as a model system for evolutionary biology. Molecular Biology and Evolution, 26(3): 623-629.
|
Yamanoue Y, Miya M, Matsuura K, Yagishita N, Mabuchi K, Sakai H, Katoh M, Nishida M. 2007. Phylogenetic position of tetraodontiform fishes within the higher teleosts:Bayesian inferences based on 44 whole mitochondrial genome sequences. Molecular Phylogenetics and Evolution, 45(1): 89-101.
|
Yu J N, Kwak M. 2015. The complete mitochondrial genome of Brachymystax lenok tsinlingensis (Salmoninae, Salmonidae) and its intraspecific variation. Gene, 573(2): 246-253.
DOI:10.1016/j.gene.2015.07.049 |
Zhang C G, Zhao Y H, Xing Y C, Zhou W, Tan, W Q. 2016.Species Diversity and Distribution of Inland Fishes in China. Science Press, Beijing. 284p. (in Chinese with English abstract)
|
Zhou L W, Feng Y, Wang G F, Wu N H. 1993. Structure analysis of the mitochondrial tRNA(Cys) gene and the original region of light-strand replication from carp. Acta Biologiae Experimentalis Sinica, 26(4): 463-468.
(in Chinese with English abstract) |
 2020, Vol. 38
2020, Vol. 38