Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LOU Yanan, SUN Bin, ZHANG Lina, LI Yong, XIAO Peng
- Establishment and characterization of a new cell line derived from half-smooth tongue sole Cynoglossus semilaevis kidney
- Journal of Oceanology and Limnology, 38(2): 560-570
- http://dx.doi.org/10.1007/s00343-019-9084-y
Article History
- Received Mar. 26, 2019
- accepted in principle May. 5, 2019
- accepted for publication Jul. 29, 2019
2 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, China;
3 University of Chinese Academy of Sciences, Beijing 100049, China;
4 Tianjin Weishan Road Middle School, Tianjin 300222, China
In vitro cell culture has gradually developed into an indispensable tool in many fields of biological research. Since the establishment of the first stable fish cell line, a gonadal cell line (RTG-2 cells) from rainbow trout (Salmo gairdneri) (Wolf and Quimby, 1962), more than 280 other fish cell lines have been established and used in many studies and applications. Cultivation of cells in vitro has promoted the advancement of knowledge in various fields, including functional genomics (Crespo et al., 2013), carcinogenesis (Lakra et al., 2011), developmental biology (Webb et al., 2015), immunology (Clem et al., 1996), virology (Gamil et al., 2015), genotoxicity (Rocco et al., 2014), gene expression (Barman et al., 2014), differentiation, developmental potentiality (Chen et al., 2010), physiology (Rode et al., 1997), and transgenic applications (Fan and Collodi, 2002). From these cell lines, in vitro experiments have identified many fish pathogens, thereby improving our understanding of fish diseases.
The half-smooth tongue sole (Cynoglossus semilaevis Günther) is a commercially valuable flatfish. Owing to its high nutritional content and appealing taste, tongue sole is one of the most important cultured fish species in China (Liao et al., 2009). However, with increased scale of aquaculture for this species, disease has become a major limitation affecting the growth of the tongue sole culture industry. Recently, more research groups have started to focus on identification of tongue sole pathogens, and several pathogens, including Vibrio harveyi (Chen, 2012), Listonella anguillarum (Zhang et al., 2009), V. rotiferianus (Chen et al., 2012), Photobacterium damselae subsp. piscicida (Wang et al., 2007), Mycobacterium marinum (Luo et al., 2018), nervous necrosis virus (NNV) (Li et al., 2014), and a novel virus (Xiao et al., 2015), have been reported to be associated with diseases in the tongue sole. Compared with diseases in other fish species, those in the tongue sole exhibit more complicated epidemiologic features, making it difficult to prevent such diseases. A 4-year monitoring study showed that some bacterial strains could be isolated from diseased tongue sole, although no effective antibiotics have been developed to control diseases in the tongue sole (data not shown). Thus, additional studies are needed to improve our understanding of tongue sole diseases, and establishment of novel cell lines from the tongue sole may facilitate the identification and detection of viral pathogens in the tongue sole. Although several cell lines have already been established from the tongue sole, including a liver cell line (HTLC cells) (Ren et al., 2008), an embryonic cell line (CFEC cells) (Sha et al., 2010), a heart cell line (CSH cells) (Wang et al., 2010), a testicular cell line (CGTC cells) (Zhang et al., 2011), a head kidney cell line (TSHKC cells) (Zheng et al., 2012), and a gonad cell line (CSPMG cells) (Sun et al., 2015), none of these cell lines have been used for identifying viral pathogens.
Therefore, in this study, we established a cell line from the tongue sole and evaluated its characteristics, with the goal of using this cell line to study viral pathogens in this fish species.
2 MATERIAL AND METHOD 2.1 Primary cell culture and subcultureSeveral healthy half-smooth tongue sole (C. semilaevis) weighing 200–250 g were obtained from New Continent Food Co., Ltd. The fishes were maintained in laboratory aquaria for at least 2 weeks. The surface of the fish was wiped with 75% v/v ethanol solution, and the kidney was removed aseptically, transferred to a dish, and washed with Dulbecco's modified Eagle's medium (DMEM)/F-12 (DMEM/F-12 powder with 4.76 g HEPES, pH 7.2) containing antibiotics (400 U/mL penicillin, 400 μg/ mL streptomycin). The kidney tissues were then washed three times with sterile phosphate-buffered saline (PBS), minced thoroughly with scissors, and digested with 1-mL trypsin solution for 10 min. Subsequently, 2-mL DMEM/F-12 complete medium (DMEM/F-12 supplemented with 20% fetal bovine serum [FBS], 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 ng/mL basic fibroblast growth factor [bFGF]) was added to the dish. The mixture of cells and cell clumps was centrifuged at 2 200 r/min for 2 min, and the pellet was suspended in 1-mL DMEM/F-12 complete medium and transferred to T-25 tissue culture flasks. The cells were incubated at 25℃ in an incubator with a normal atmosphere. The next day, 1-mL DMEM/F-12 complete medium was added to the flask. One half of the DMEM/F-12 complete medium was changed every 3 days.
When the cells around the tissue pieces formed a monolayer, the old medium was removed, and the cells were washed with PBS. Cell clusters were digested with 1-mL trypsin and cultured in the original culture flask with 2-mL DMEM/F-12 complete medium. Dissociation was monitored under an inverted light microscope (Nikon TE2000-S) to ensure that cells had been released from the flask surface. For 10 passages, the cells were cultured in the original culture flask. After 10 passages, cells were subcultured and transferred to new T-25 flasks at a split ratio of 1:2. The primary cells were initially maintained in DMEM/F-12 complete medium with 20% FBS.
2.2 Growth curveFor the growth study, the cells at passage 18 were seeded into 12-well plates at an initial density of 2×105 cells/mL and incubated at 25℃ for 8 days. One well of cells was digested each day with 0.25% trypsin, and the number of cells was counted using a hemocytometer. The experiment was performed in triplicate. For construction of cell growth curves, the cultivation time was set as the abscissa and the number of cells per milliliter was set as the ordinate.
2.3 Effects of temperature on cell proliferationTo analyze how temperature affected the growth of the cells, 2×104 cells/mL at passage 22 were inoculated in three 12-well plates and incubated at 16℃, 25℃, or 30℃. Cell numbers were counted microscopically using a hemocytometer every day for 8 days. The experiment was performed in triplicate.
2.4 Chromosome analysisFor chromosome analysis, we used CSK cells at passage 23. Briefly, the cells were seeded in T-25 flasks and cultured at 25℃ overnight. The cells were then treated with colchicine (1 μg/mL) for 3.5 h at 25℃. The old medium was removed, cells were trypsinized, and DMEM/F-12 complete medium was added to the flasks. The cells were harvested by centrifugation (650×g, 3 min). The pellets were resuspended in 5-mL hypotonic solution (0.075 mol/L KCl) and incubated at 37℃ for 30 min. The cells were then fixed for 2 min in 1-mL precooled Carnoy's fixative (methanol:acetic acid=3:1), centrifuged at 1 000 r/min for 6 min, fixed twice in 2-mL precooled Carnoy's fixative (15 min each time), centrifuged again, and resuspended in 0.5-mL precooled Carnoy's fixative. Finally, slides were prepared using the conventional drop splash technique (Freshney) and then air dried. Chromosomes were stained with 5% Giemsa for 30 min. One hundred photographed cells at metaphase were counted under a Nikon eclipse 80-I fluorescence microscope, and chromosome karyotype was analyzed according to the method of Levan et al. (1964).
2.5 Sex genotyping of CSK cellsA DNA extraction kit was used to extract DNA from the established half-smooth tongue sole kidney cell line. Briefly, based on sex-specific sequences, we designed primers for the sequence characterized amplified region, as follows: sense primer, 5′-CCTAAATGATGGATGTAGATTCTGTC-3′, and reverse primer, 5′-GATCCAGAGAAAATAAACCCAGG-3′ (Liu et al., 2014). In females, these primers yield two DNA fragments of 169 and 134 bp, whereas in males, these primers yield one DNA fragment of 169 bp. The polymerase chain reaction (PCR) conditions were as follows: initial denaturation at 95℃ for 10 min; 35 cycles of 95℃ for 30 s, 54℃ for 30 s, and 72℃ for 40 s; and termination of the reaction at 72℃ for 10 min. The PCR products (8 μL) were resolved on 2% agarose gels with a Trans 2k plusⅡDNA marker.
2.6 Virus susceptibilityCSK cells were used to detect susceptibility to Grouper NNV. Briefly, we seeded CSK cells at passage 26 into T-25 flasks and incubated the cells overnight. When the cell confluence was approximately 90%, the medium was removed, and the cells were washed twice with PBS. Next, 1-mL NNV solution diluted with DMEM/F-12 minimal medium was added to the flasks. After 1 h, the virus solution was replaced with 3-mL DMEM/F-12 complete medium containing 5% FBS. The cytopathic effect (CPE) was observed every 12 h using an inverted microscope. For observation of the CPE, we collected NNV-infected cells, fixed the cells with 2.5% glutaraldehyde in cacodylate buffer (0.1 mol/L, pH 7.4) for 4 h at 4℃, rinsed the cells three times with PBS (0.1 mol/L, pH 7.4), and postfixed the cells with 1% osmium tetroxide in cacodylate buffer (0.1 mol/L, pH 7.4) for 2 h. The samples were then rinsed three times in PBS buffer for 10 min. We used graded ethyl alcohol (30%, 50%, 70%, 90%, and 100%) to dehydrate the samples and then embedded the samples in Epon8-12 epoxy resin. A diamond knife was used to cut ultrathin sections on a ReichertJung Ultracut-E microtome. Samples were then mounted on copper grids, stained with 2% uranyl acetate-lead citrate, and examined with a JEOL JEM- 1200EX transmission electron microscope. NNVinfected cell supernatants were harvested and fixed with 2.5% glutaraldehyde in PBS (0.1 mol/L, pH 7.4) for 15 min. Subsequently, the samples were mounted on copper grids, stained with 3% phosphotungstic acid, and washed with double distilled water three times. The copper grids were placed in a desiccator and then examined with a JEOL JEM-1200EX transmission electron microscope.
2.7 Cell transfection with the pEGFP-N1 reporter geneCSK cells were seeded at a density of 1×106 cells/ well in 12-well plates and incubated at 25℃. When the cells reached 85%–90% confluence, the cells were transfected with the pEGFP-N1 expression vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Briefly, 2-μL Lipofectamine 2000 was mixed with 98-μL Opti-MEM (Invitrogen) in a 0.5-mL sterile centrifuge tube. At the same time, 2-μL pEGFP-N1 (500 ng/μL) was mixed with 98-μL OptiMEM (Invitrogen) in a 0.5-mL sterile centrifuge tube. After 5 min, the two solutions were mixed and then incubated together for 20 min. The cells were washed twice with PBS, and then the medium was substituted with Opti-MEM. The mixtures were then dropped into the wells, and cells were cultured at 25℃ for 5.5 h. The medium was then replaced with normal medium. After incubation for 24 h, the cells were washed twice with PBS, fixed with 4% paraformaldehyde (PFA) for 10 min, and washed twice with PBS. The nuclei were then stained with 4, 6-diamino-2-phenylindole (DAPI) for 15 min and washed with PBS twice. Finally, the green and blue fluorescence signals were observed under afl uorescence microscope (Zeiss LSM710).
2.8 Cryopreservation and recovery of cellsCells at 85%–90% confluence were used for cryopreservation. The cells were trypsinized and suspended in DMEM/F-12 complete medium and then centrifuged at 2 200 r/min for 2 min. The cell pellet was resuspended with the medium containing 10% dimethyl sulfoxide and 90% FBS. Cells were then dispensed into 2-mL sterile plastic tubes, kept in a NALGEBE Cryo 1℃ Freezing Container (Invitrogen) at -80℃ overnight, and transferred to liquid nitrogen (-196℃) the next day. When thawed, the cryopreserved cells were placed into double distilled water at 42℃ until just thawed and then centrifuged at 2 200 r/min for 2 min. The cells were suspended in DMEM/F-12 complete medium, transferred to T-25 flasks, and cultured at 25℃ in a normal atmosphere incubator.
2.9 ImmunocytochemistryAt passage 37, CSK cells were used for immunocytochemical analysis with specific antibodies for supporting cells. The cells were seeded at a density of 1×105 cells/well in 12-well plates and cultured at 25℃. When the monolayer reached 75%– 85% confluence, the old medium was removed, and the cells were washed with precooled PBS twice and fixed with 4% PFA. The cells were washed with PBS and perforated with 0.2% TritonX-100 for 15 min. The cells were then washed with PBS twice, blocked with 1% bovine serum albumin for 30 min, and incubated with primary antibodies (Table 1) at 4℃ overnight. After incubation, the cells were washed with PBS twice and then incubated again with secondary antibodies (Alexa-Fluor 488-labeled Goat Anti-Rabbit IgG (H+L) and Alexa-Fluor 488-labeled Goat Anti-Mouse IgG (H+L)) in the dark for 1.5 h at room temperature. Subsequently, the cells were washed with PBS twice and stained with DAPI for 15 min. A fluorescence microscope (Zeiss LSM710) was then used to observe the results. For the negative control, the primary antibodies were omitted.
Approximately 3–5 days after the tissue pieces and cell suspensions were inoculated onto the plates, the cells began to move out from the edges of the tissue blocks. When the cells grew to 90% confluence, the first subculture was performed. After 10 passages, the cells were subcultured at a ratio of 1:2 for (3–7)-day intervals. The culture time for cells at early passages was longer than that for later cells. The cells showed a fibroblast-like phenotype (Fig. 1).

|
| Fig.1 Morphology characterization of the CSK cells a. primary culture on the 4th day following seeding of tissue blocks, bar=50 μm; b. at passage 3, bar=50 μm; c. at passage 56, bar=25 μm. |
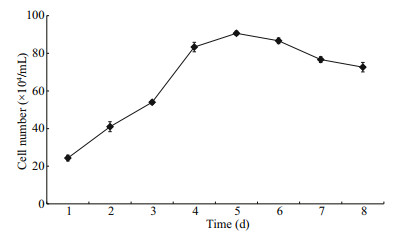
Cells were plated at a density of 2×105 and incubated for 24 h. The growth curve of CSK cells is shown in Fig. 2. After 4 days, the cell number reached 8.9×105 cells/mL.
|
| Fig.2 The CSK cells growth curve |
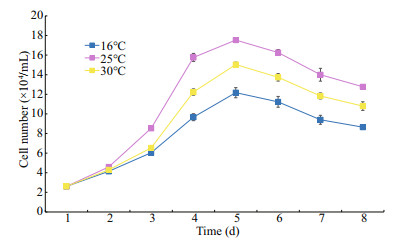
At passage 22, CSK cells were able to grow at temperatures ranging from 16 to 30℃. The cell numbers were determined at different temperatures for 8 consecutive days. The highest growth rate was obtained at 25℃, with cell numbers reaching 17.45× 104 cells/mL after 4 days, and the lowest growth rate was obtained at 16℃, with the cell number reaching only 12.17×104 cells/mL (Fig. 3). There were no obvious differences in the morphological characteristics of cells cultured under different temperatures.
|
| Fig.3 Effects of temperature on the growth of the CSK cells |
Chromosome analysis of cells at passage 23 showed that there was a chromosome number of 42 for 43% of 100 metaphase cells (Fig. 4c). During metaphase (Fig. 4a), cells having a normal diploid number also exhibited a normal karyotype morphology (Fig. 4b).

|
| Fig.4 Chromosome analysis of CSK cells at passage 23 a. metaphase; b. diploid karyotype; c. chromosome number distribution, one hundred metaphases were counted. |
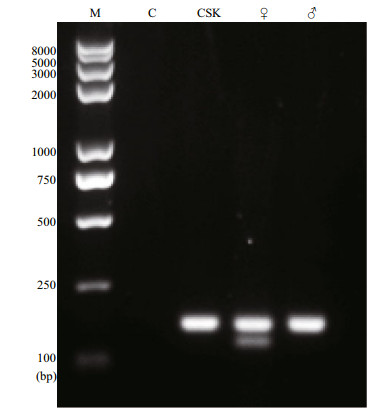
The results revealed that the cells had a diploid karyotype consisting of 21 pairs of chromosomes; the female-specific W chromosome was absent (Fig. 4a). Accordingly, we concluded that CSK cells were male cells. Furthermore, PCR analysis revealed that the CSK cells yielded only the 169-bp male DNA fragment, further confirming that these cells had been derived from a male fish (Fig. 5).
|
| Fig.5 Half-smooth tongue sole sex specific markers to identify CSK cells M: trans 2k plusⅡ DNA marker; C: no DNA temple control; CSK: identified CSK cells; ♀: female positive control; ♂: male positive control. |
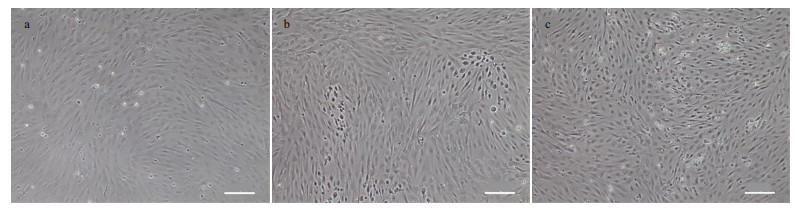
CSK cells were susceptible to NNV, and a CPE was observed at 3 days after NNV infection. The cells were shrunk and became circular, and the cell nucleus was black and dense (Fig. 6).
|
| Fig.6 NNV-infected cells a. uninfected cells, bar=100 μm; b–c. the CPE after 3 days after infection NNV, bar=100 μm. |
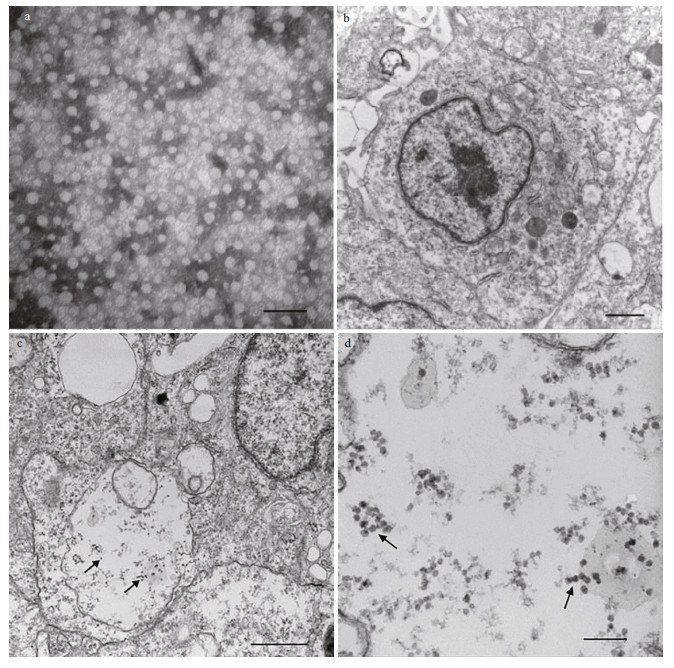
When we observed a typical CPE, the cells were collected for TEM analysis. We found scattered virus particles in the cytoplasm (Fig. 7b–d) and changes in the cell morphology after virus infection, suggesting that the CSK cells were sensitive to NNV.
|
| Fig.7 NNV infected CSK cell a. electron micrograph of NNV particles before infection, bar=100 nm; b. uninfected CSK cells, bar=1μm; c. electron micrograph of NNV-infected CSK cells, bar=1μm; d. the local amplification of cytoplasm include NNV, bar=200 nm. |
The cells were successfully transfected with pEGFP-N1 using Lipofectamine 2000, and clear, strong green fluorescence signals were detected after 24 h (Fig. 8). The efficiency of transfection was about 26.9%, suggesting that CSK cells were suitable for transfection.

|
| Fig.8 The expression of pEGFP-N1 in CSK cells a. control CSK cells dyed DAPI; b. transfected cells; c. mixture of a and b. Bar=20 μm. |
CSK cells were cryopreserved at different passages. After thawing and seeding into flasks, the cells recovered with a survival rate of 70%–80%. The cells could grow to confluence after 7 days.
3.9 ImmunocytochemistryAntibodies specific for epithelial cells and stromal cells were used for immunocytochemical analysis in CSK cells. RLT1572 detects endogenous epithelial cell adhesion molecule protein. RLT5053 and RLM3364 detect endogenous α-SMA protein. RLT1453 detects endogenous E-cadherin protein. RLT1275, RLM3095, and RLM3080 detect endogenous different Cytokeratin proteins. The results showed that most cells presented fluorescence signal, which means CSK cells were mainly supporting cells (Fig. 9).

|
| Fig.9 Immunocytochemical analysis of CSK cells by specific antibodies The cells were present as filamentous cells, bar=20 μm. a. RLT1572; b. RLT5053; c. RLT1453; d. RLT 1275; e. RLM 3095; f. RLM3364; g. RLM 3080. |
With the expansion of aquaculture scale owing to the global demand for aquaculture products, the development of the aquaculture industry is facing increasing problems, particularly with regard to the frequency of new diseases. New sequencing technologies have detected novel viruses in diseased organisms. However, we have not obtained direct evidence demonstrating that these viruses are the cause of disease. Cell culture is an essential tool for biological research and has facilitated the identification of many viral pathogens from various hosts (John and Richards, 1999; Isshiki et al., 2004). Notably, in previous studies, many bacterial pathogens have been isolated from diseased tongue sole, although no effective antibiotics have been developed to control these diseases. Thus, we still have an incomplete understanding of tongue sole diseases, and additional cell lines are urgently needed to support further research in this fish species.
In this study, a half-smooth tongue sole (C. semilaevis) kidney cell line was successfully developed and characterized, designated as CSK cells. We characterized the growth, transfection efficiency, NNV susceptibility, and internal and external morphologies of the CSK cell line. Our results showed that CSK cells grew stably in DMEM/F-12 supplemented with FBS and bFGF. The cells were subcultured for 100 passages and showed a fibroblasticlike morphology.
CSK cells could grow under different temperatures ranging from 16 to 30℃, with optimal growth at 25℃. Notably, this optimum temperature was the temperature at which the primary culture was established and was similar to that of other halfsmooth tongue sole cell lines (Ren et al., 2008; Sha et al., 2010; Wang et al., 2010; Zhang et al., 2011; Sun et al., 2015). These results also demonstrated that fish cell culture is different from mammalian cell culture, and fish cells showed many advantages over mammalian cells (Bols and Lee, 1991). For example, fish cells can be cultured at room temperature (16– 30℃) and can be directly exposed to environment of the same atmospheric pressure. Additionally, fish cells that adapt to a wide temperature range can be used for a wider range of applications, suggesting potential suitability for isolating both warm water and cold water fish viruses (Nicholson et al., 1987).
From the well-spread metaphase chromosomes obtained in this study, we concluded that C. semilaevis had a diploid chromosome number of 42 (Shao et al., 2010). Indeed, karyotype analysis revealed that 43% of the CSK cells possessed a diploid chromosome number of 2n=42, but lacked the heterogametic W chromosome. Furthermore, PCR analysis revealed that the CSK cell line did not have a female-specific marker, exhibiting the ZZ genetic constitution.
The susceptibility of CSK cells to NNV was evaluated by observation of the CPE. At 3 days after infection, cells inoculated with NNV exhibited morphological changes, including cell shrinkage and black and dense nuclei. Electron microscopy observations revealed that some virus particles were scattered throughout the cytoplasm of cells infected with NNV. As a viral pathogen to tongue sole (Li et al., 2014), NNV is also a causal agent of disease in many fish species. Although some cell lines have been developed for reproduction of NNV (Chi et al., 2005), development of this cell line from tongue sole will provide more appropriate conditions for studies of viruses. The results showed that the CSK cell line was susceptible to NNV, providing better condition for virus research.
Kidneys are comprised of many specialized cells, which facilitate diverse activities. Renal cells are broadly classified as parenchymal (functional cells) or stromal cells (support cells), and these categories include diverse epithelial and mesenchymal cell types. The epithelial cells are arranged into working units called nephrons, and the mesenchymal cells occupy the intervening interstitial spaces between nephrons (Gerlach and Wingert, 2013). Our findings demonstrated that CSK cells were mainly support cells.
In conclusion, we established a new cell line from the kidney of the half-smooth tongue sole. This cell line is expected to provide a useful tool for the identification of viral infections and for exogenous gene expression in the half-smooth tongue sole. Furthermore, this cell line may be instrumental in the analysis of mutant gene function in the context of kidney disease at a cellular level.
5 DATA AVAILABILITY STATEMENTAll data generated and/or analyzed during this study are available from the corresponding author on reasonable request.
Barman A S, Lal K K, Rathore G, Mohindra V, Singh R K, Singh A, Khare P, Lal B. 2014. Derivation and characterization of a es-like cell line from indian catfish heteropneustes fossilis blastulas. Scientific World Journal, 2014: 427497.
|
Bols N C, Lee L E J. 1991. Technology and uses of cell cultures from the tissues and organs of bony fish. Cytotechnology, 6(3): 163-187.
DOI:10.1007/BF00624756 |
Chen H, Yi Y, Chen M, Yang X. 2010. Studies on the developmental potentiality of cultured cell nuclei of fish. International Journal of Biological Sciences, 6(2): 192-198.
|
Chen J. 2012. Bacterial Diseases and Control of Tongue Fish(Cynoglossus semilaevis) in Recirculating Culture System. Shanghai Ocean University, Shanghai. 85p. (in Chinese with English abstract)
|
Chen Z Q, Yao Z X, Lin M, Chang J B. 2012. Characterization of pathogenic bacteria Vibrio rotiferianus isolated from Cynoglossus semilaevis Günther. Biotechnology Bulletin, (6): 147-153.
(in Chinese with English abstract) |
Chi S C, Wu Y C, Cheng T M. 2005. Persistent infection of betanodavirus in a novel cell line derived from the brain tissue of barramundi Lates calcarifer. Diseases of Aquatic Organisms, 65(2): 91-98.
|
Clem L W, Bly J E, Wilson M, Chinchar V G, Stuge T, Barker K, Luft C, Rycyzyn M, Hogan R J, Van Lopik T, Miller N W. 1996. Fish immunology, the utility of immortalized lymphoid cells-a mini review. Veterinary Immunology and Immunopathology, 54(1-4): 137-144.
DOI:10.1016/S0165-2427(96)05682-6 |
Crespo B, Zanuy S, Gómez A. 2013. Development of an in vitro system for functional studies of ovarian follicular cells in European sea bass (Dicentrarchus labrax). Cytotechnology, 65(2): 273-286.
DOI:10.1007/s10616-012-9484-8 |
Fan L C, Collodi P. 2002. Progress towards cell-mediated gene transfer in zebrafish. Briefings in Functional Genomics & Proteomics, 1(2): 131-138.
|
Freshney R I. 2010. Culture of Animal Cells. 6th edn. New York: John Wiley& Sons, p.239-268.
|
Gamil A A A, Evensen Ø, Mutoloki S. 2015. Infection profiles of selected aquabirnavirus isolates in CHSE cells. PLoS One, 10(8): e134173.
|
Gerlach G F, Wingert R A. 2013. Kidney organogenesis in the zebrafish:insights into vertebrate nephrogenesis and regeneration. Wiley Interdisciplinary Reviews:Developmental Biology, 2(5): 559-585.
DOI:10.1002/wdev.92 |
Isshiki T, Nagano T, Kanehira K, Suzuki S. 2004. Distribution of marine birnavirus in cultured marine fish species from Kagawa Prefecture, Japan. Journal of Fish Diseases, 27(2): 89-98.
DOI:10.1111/j.1365-2761.2003.00518.x |
John K R, Richards R H. 1999. Characteristics of a new birnavirus associated with a warm-water fish cell line. Journal of General Virology, 80(8): 2 061-2 065.
DOI:10.1099/0022-1317-80-8-2061 |
Lakra W S, Swaminathan T R, Joy K P. 2011. Development, characterization, conservation and storage of fish cell lines:a review. Fish Physiology and Biochemistry, 37(1): 1-20.
|
Levan A, Fredga K, Sandberg A A. 1964. Nomenclature for centromeric position on chromosomes. Hereditas, 52(2): 201-220.
|
Li J, Shi C, Huang J, Geng W G, Wang S Q, Su Z D. 2014. Complete genome sequence of a betanodavirus isolated from half-smooth tongue sole (Cynoglossus semilaevis). Genome Announcements, 2(6): e01312-14.
|
Liao X L, Ma H Y, Xu G B, Shao C W, Tian Y S, Ji X S, Yang J F, Chen S L. 2009. Construction of a genetic linkage map and mapping of a female-specific DNA marker in half-smooth tongue sole (Cynoglossus semilaevis). Marine Biotechnology, 11(6): 699-709.
DOI:10.1007/s10126-009-9184-3 |
Liu Y, Chen S L, Gao F T, Meng L, Hu Q M, Song W T, Shao C W, Lv W Q. 2014. SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole(Cynoglossus semiliaevis). Journal of Agricultural Biotechnology, 22(6): 787-792.
(in Chinese with English abstract) |
Luo Z, Li J, Zhang Z G, Hao S, Bai X H, You H Z, Mo Z L, Feng S M. 2018. Mycobacterium marinum is the causative agent of splenic and renal granulomas in half-smooth tongue sole (Cynoglossus semilaevis Günther) in China.Aquaculture, 490: 203-207.
|
Nicholson B L, Danner D J, Wu J L. 1987. Three new continuous cell lines from marine fishes of Asia. In Vitro Cellular & Developmental Biology, 23(3): 199-204.
|
Ren G C, Chen S, Sha Z X. 2008. Development and characterization of a liver cell line from half-smooth tongue-sole (Cynoglossus semilaevis). Chinese High Technology Letters, 18(6): 657-660.
(in Chinese with English abstract) |
Rocco L, Valentino I V, Scapigliati G, Stingo V. 2014. RAPDPCR analysis for molecular characterization and genotoxic studies of a new marine fish cell line derived from Dicentrarchus labrax. Cytotechnology, 66(3): 383-393.
DOI:10.1007/s10616-013-9586-y |
Rode M, Berg T, Gjøen T. 1997. Effect of temperature on endocytosis and intracellular transport in the cell line SHK-1 derived from salmon head kidney. Comparative Biochemistry and Physiology Part A:Physiology, 117(4): 531-537.
DOI:10.1016/S0300-9629(96)00414-8 |
Sha Z X, Ren G C, Wang X L, Wang N, Chen S L. 2010. Development and characterization of a cell line from the embryos of half smooth tongue sole (Cynoglossus semilaevis). Acta Oceanologica Sinica, 29(2): 81-87.
DOI:10.1007/s13131-010-0025-z |
Shao C W, Wu P F, Wang X L, Tian Y S, Chen S L. 2010. Comparison of chromosome preparation methods for the different developmental stages of the half-smooth tongue sole, Cynoglossus semilaevis. Micron, 41(1): 47-50.
|
Sun A, Chen S L, Gao F T, Li H L, Liu X F, Wang N, Sha Z X. 2015. Establishment and characterization of a gonad cell line from half-smooth tongue sole Cynoglossus semilaevis pseudomale. Fish Physiology and Biochemistry, 41(3): 673-683.
DOI:10.1007/s10695-015-0037-x |
Wang X L, Wang N, Sha Z X, Chen S L. 2010. Establishment, characterization of a new cell line from heart of half smooth tongue sole (Cynoglossus semilaevis). Fish Physiology and Biochemistry, 36(4): 1 181-1 189.
DOI:10.1007/s10695-010-9396-5 |
Wang Y, Han Y, Li Y, Chen J X, Zhang X H. 2007. Isolation of photobacterium damselae subsp. piscicida from diseased tongue sole (Cynoglossus semilaevis Günther) in China.Acta Microbiologica Sinica, 47(5): 763-768.
(in Chinese with English abstract) |
Webb C F, Ratliff M L, Powell R, Wirsig-Wiechmann C R, Lakiza O, Obara T. 2015. A developmentally plastic adult mouse kidney cell line spontaneously generates multiple adult kidney structures. Biochemical and Biophysical Research Communications, 463(4): 1 334-1 340.
DOI:10.1016/j.bbrc.2015.06.130 |
Wolf K, Quimby M C. 1962. Established eurythermic line of fish cells in vitro. Science, 135(3508): 1 065-1 066.
DOI:10.1126/science.135.3508.1065 |
Xiao P, Jiang M, Liu Y, Sun M, Zhang L, Jie L, Li G, Mo Z. 2015. Splenic necrosis signs and pathogen detection in cultured half-smooth tongue sole, Cynoglossus semilaevis Günther. Journal of Fish Diseases, 38(1): 103-106.
DOI:10.1111/jfd.12213 |
Zhang B, Wang X L, Sha Z X, Yang C G, Liu S S, Wang N, Chen S L. 2011. Establishment and characterization of a testicular cell line from the half-smooth tongue sole, Cynoglossus semilaevis. International Journal of Biological Sciences, 7(4): 452-459.
DOI:10.7150/ijbs.7.452 |
Zhang X J, Qin G M, Yan B L, Xu J, Bi K R, Qin L. 2009. Phenotypic and molecular characterization of pathogenic Listonella anguillarum isolated from half-smooth tongue sole Cynoglossus semilaevis. Acta Oceanologica Sinica, 31(5): 112-122.
(in Chinese with English abstract) |
Zheng Y, Wang N, Xie M S, Sha Z X, Chen S L. 2012. Establishment and characterization of a new fish cell line from head kidney of half-smooth tongue sole (Cynoglossus semilaevis). Fish Physiology and Biochemistry, 38(6): 1 635-1 643.
DOI:10.1007/s10695-012-9660-y |
 2020, Vol. 38
2020, Vol. 38