Institute of Oceanology, Chinese Academy of Sciences
Article Information
- HONG Pei, GONG Shihao, WANG Chunbo, SHU Yilin, WU Xingqiang, TIAN Cuicui, Oscar Omondi DONDE, CAI Pei, WU Huaming, XIAO Bangding
- Effects of organic carbon consumption on denitrifier community composition and diversity along dissolved oxygen vertical profiles in lake sediment surface
- Journal of Oceanology and Limnology, 38(3): 733-744
- http://dx.doi.org/10.1007/s00343-019-9103-z
Article History
- Received Apr. 18, 2019
- accepted in principle Jul. 24, 2019
- accepted for publication Aug. 19, 2019
2 University of Chinese Academy of Sciences, Beijing 100049, China;
3 College of Life Sciences, Anhui Normal University, Wuhu 241000, China
Nitrate pollution in both surface and groundwater has for a long time been one of the most important water quality issues worldwide (Nolan, 2001; Puckett et al., 2011). Sediment denitrification is the main process of nitrogen removal in shallow lakes (Sirivedhin and Gray, 2006). Normally, reduced oxygen concentrations and the availability of electron donors (i.e. organic carbon) and N sources were the three conditions necessary for the denitrification. In nature, the organic carbon compounds are the most common carbon source for the denitrification process (Van Rijn et al., 2006) and the nature of organic carbon has influence on the denitrification performance, microbial community structure as well as denitrification genes (Srinandan et al., 2012; Xu et al., 2018). Previous studies have reported on the effects of organic matter on denitrification in soil and sediments of the lakes, oceans and rivers (Groffman et al., 2009; Huang et al., 2011; Jenerette and Chatterjee, 2012; Wu et al., 2013; Jia et al., 2016; Liu et al., 2018a). Dissolved oxygen (DO) is usually considered the most critical proximal regulator of microbial denitrification (Cao et al., 2017). Different denitrifying enzymes have varying sensitivity to different oxygen concentrations (McKenney et al., 2001). Nitrogen removal in wastewater treatment systems with different DO concentrations has been significantly studied (Chen et al., 2016; Waki et al., 2018; Zou et al., 2018), but the response of denitrifying bacteria is still not adequately reported. Previous studies on the physical and chemical properties of the lake sediment-water interface found that the surface sediments had severe redox gradient changes within the range of several millimeters (Santschi et al., 1990). However, under condition of continuous degradation of organic matter, the characteristics of denitrifying bacteria in surface sediments on the vertical scale based on changes of DO content are still not adequately known.
Various reductases catalyzed nitrate to nitrite, nitric oxide, nitrous oxide and dinitrogen gas through the process of denitrification (Zumft, 1997). Through this process, nirS gene that encodes for cytochrome-cd1 nitrite reductase, catalyzes the first step in the evolution of the gaseous product (NO2--NO). The nirS gene is more widely distributed amongst the bacteria from the natural environments (Braker et al., 1998) and has been most frequently used as functional biomarkers of the denitrifying community (Bulow et al., 2008; Francis et al., 2013; Yang et al., 2013; Gao et al., 2016).
In the current study, three lake sediment layers: aerobic zone (AEZ), hypoxic-anoxic zone (HAZ) and anoxic zone (ANZ), distinguished using an oxygen microsensor on a vertical scale over organic carbon consumption rate, were stratified and sampled. The aim of this study was to explore the dynamics in denitrifying bacteria abundance, community composition, structure, and diversity along the vertical DO boundary within the surface sediment profiles using nirS gene. This was to provide a deeper understanding on the traits of denitrifying bacteria based on the consumption of organic carbon within a eutrophic lake. The information generated from this study is helpful in providing accurate understanding on the contribution of lake surface sediments to denitrification process, providing insights necessary in developing eutrophication control strategies.
2 MATERIAL AND METHOD 2.1 Experimental designSurface sediment was collected in May 2018 from Dianchi Lake, located in Kunming, China (102°38ʹ0.84ʹʹE, 24°58ʹ41.61ʹʹN) using specific sampling methods described previously by Tian et al. (2015). Surface sediment cores were sampled to a depth of 20 mm and carefully transferred to PVC cylinders in situ (30 mm diameter × 110 mm high) before being subject to laboratory microcosm incubations. A simulation of the overlying lake water contained in mg/L; 48.6 NaNO3, 5.1 MgSO4·7H2O, 3.8 NH4Cl, 5.6 K2HPO4, 4.4 KH2PO4, 0.1 mL/L of trace elements (Nancharaiah et al., 2008) and pH of 7.2. A 20-mL aliquot of synthetic lake water was then gently overlaid onto the sediment using a siphon to avoid disturbance. The depth of the added synthetic lake water was approximately 30 mm above the sediment, resulting in a water׃sediment ratio as described in Rong et al. (2016). The nitrate concentration in the overlaid water was measured 1to 2 times per day with an ion chromatography (ICS5000þ, Thermo Fisher Scientific, MA, USA), and when it was lower than 1 mg/L, the water was replaced. All cylinders were stored in dark incubators at 25℃ for 30 days. Sampling for denitrifier bacterial richness was set up at day 10 when the organic carbon content was high and at day 30 when the organic carbon content was low.
2.2 Sediment analysesAt days 10 and 30, the sediment was acquired and parameters measured in triplicate. Micro-sensors with a 90–110 μm tip diameter were used to detect DO levels (Unisense, Denmark). The sediment cores were cut into 3 parts of the following intervals; 0–2 mm, 2–4 mm, 4–6 mm. Frozen dried sediments were sieved to measure the total organic carbon (TOC) by an elementar vario TOC system (Elementar, Germany). Dissolved organic carbon (DOC) was extracted by water at a ratio of 1:10 (sediment/water) and measured using a DRB200 digital reactor (HACA, USA).
2.3 DNA extractionDNA was extracted from approximately 0.8 g of each sediment sample using an EZNA Soil DNA Kit (Omega, USA) in line with instructions of the manufacturer. DNA concentration and quality were assessed by spectrophotometry and 1% agarose gel electrophoresis, respectively. All DNA extracts were stored at -20℃ until further processing.
2.4 qPCR and high-throughput sequencingQuantitative PCR analysis was undertaken for the denitrification bacterial abundance based on the quantification of nirS gene and the primer pair nirScd3Af and nirSR3cd (Kandeler et al., 2006), and 16S rRNA gene with the primer pair 338F and 806R (Xu et al., 2016). Their abundance was determined by the qPCR standard curves (Bio-Rad, USA) using SYBR Green as the signal dye. Standard curve was obtained by tenfold serial dilution of standard plasmids containing target functional gene. Each 20- μL reaction mixture contained 1 μL of template DNA, 10 μL of iTaq Universal SYBR Green Supermix (BioRad), 1 μL of each primer, and 7 μL of water. The data were analyzed using the Bio-Rad software. Plasmid containing target gene and nuclease-free water as positive and negative controls respectively were run together with each sample.
The primer nirScd3aF/nirSR3cd of nirS was also used for sequencing amplification on an Illumina MiSeq platform (Illumina, USA) by LC-Bio Technology Co. (Hangzhou, China). Raw fastq files were demultiplexed and quality-filtered with Trimmomatic and merged with FLASH. Sequences with 97% similarity were clustered as Operational Taxonomic units (OTUs) with UCHIME software (version 7.1 http://drive5.com/uparse/). Taxonomy of nirS gene was analyzed using the RDP Classifier (http://rdp.cme.msu.edu/).
2.5 Statistical analysisAlpha diversity was applied in analyzing the complexity of species diversity of the samples using the Observation and Chao indices with Mothur (version v.1.30.1 http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity). Beta diversity analysis was used to evaluate differences in species complexity among the samples. Beta diversity was calculated through Analysis of Similarities (ANOSIM) with weighted Non-metric Multidimensional Scaling analysis (NMDS) in the R "vegan" package (v3.2.3). One-way and two-way ANOVA, and Pearson's correlation analysis were conducted using SPSS version 19.0 software. Other figures were drawn using the Origin 2017 program.
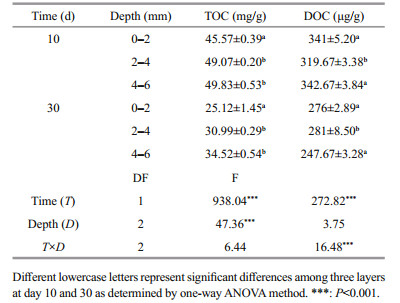
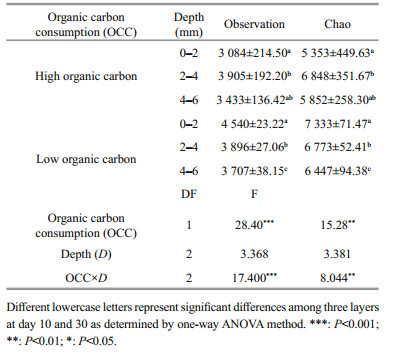
3 RESULT 3.1 Chemical properties of surface sediments among vertical DO profilesOxygen penetration depth is a measure of infiltration depth of oxygen from the water into the sediment and determines the thickness of the aerobic sediment. The experimental results revealed that oxygen penetrated into the sediment, and that the depth of penetration for oxygen concentration of 0.5 mg/L varied within the range of 2.0–2.5 mm during the 10th and 30th sampling days (Fig. 1). Based on the DO penetration depth data, it was determined that 0–2 mm, 2–4 mm, and 4–6 mm corresponded to the aerobic, hypoxia-anoxic and anoxic DO layers, respectively. Two-way ANOVA showed that TOC content (P < 0.01) and DOC content (P < 0.01) were significantly consumed over time at the three layers. Similarly, TOC content (P < 0.01) was found to be significantly different between the depths in the surface sediments (two-way ANOVA, Table 1). Furthermore, interactions between sampling days and depth were found in DOC content (P < 0.01).
|
| Fig.1 Dissolved oxygen profiles at the sediment-water interface on days 10 and 30 AEZ, HAZ and ANZ represent the aerobic, hypoxic-anoxic and anoxic DO layers, respectively. |
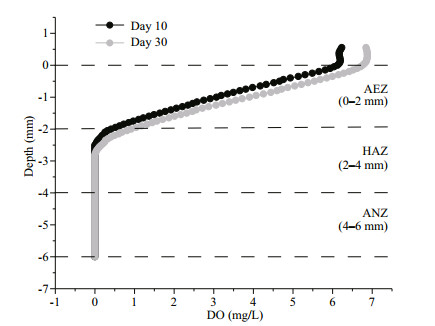
The qPCR analysis of 16S rRNA and nirS gene copies revealed that the abundance of total bacteria decreased gradually with increasing depth, while the abundance and relative abundance of nirS gene increased to a peak at hypoxia-anoxic DO layers (2–4 mm) both at high and low organic carbon sediment content (Fig. 2). Moreover, two-way ANOVA showed that the abundance of total bacteria as well as the abundance and relative abundance (ratio of nirS/16S rRNA gene copies) of nirS gene were significantly different based on the consumption of organic carbon and increase in the depth (P < 0.01; Fig. 2a, b, c). Additionally, the interaction of depth and organic carbon consumption through the abundance and relative abundance of nirS gene was significantly different (P < 0.05). The number of nirS gene sequences based on Illumina Miseq-based sequencing also showed the same trend (Fig. 2d). Moreover, Pearson's correlation showed that TOC and DOC contents were the significant factors correlating with the abundance and relative abundance of nirS gene (Fig. 2e).
|
| Fig.2 The values of 16S rRNA bacterial gene (a), the denitrifier nirS gene abundance (b), the ratio of nirS gene to 16S rRNA gene according to qPCR (c), the number of nirS gene sequences based on high-throughput sequencing (d), along the aerobic (AEZ), hypoxic-anoxic (HAZ) and anoxic (ANZ) layers on high and low organic carbon sediment surface Error bars represent standard error (n=3). The black and white square represent high organic carbon (HOC) and low organic carbon (LOC), respectively. The following table shows the results of the two-way ANOVA data obtained from the attached figures. The data in the table presented the F value (e). Pearson's correlation between organic carbon (TOC and DOC contents) and the abundance and relative abundance of nirS gene. |
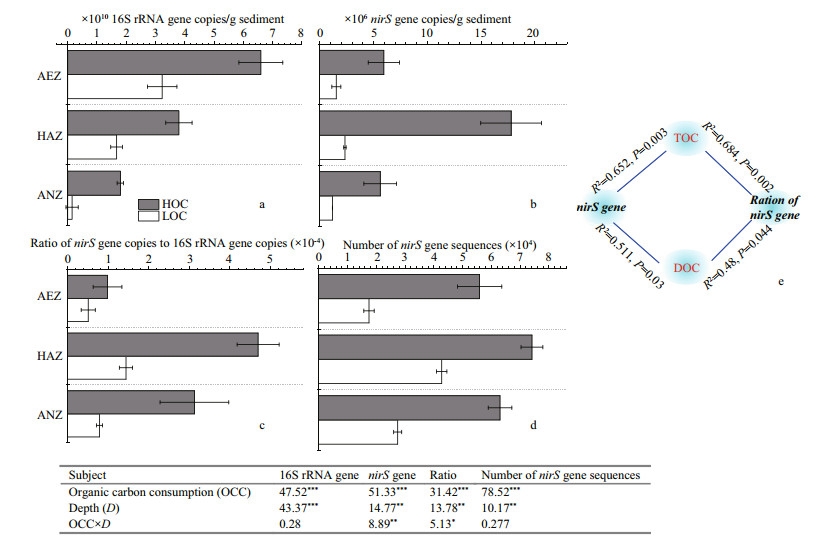
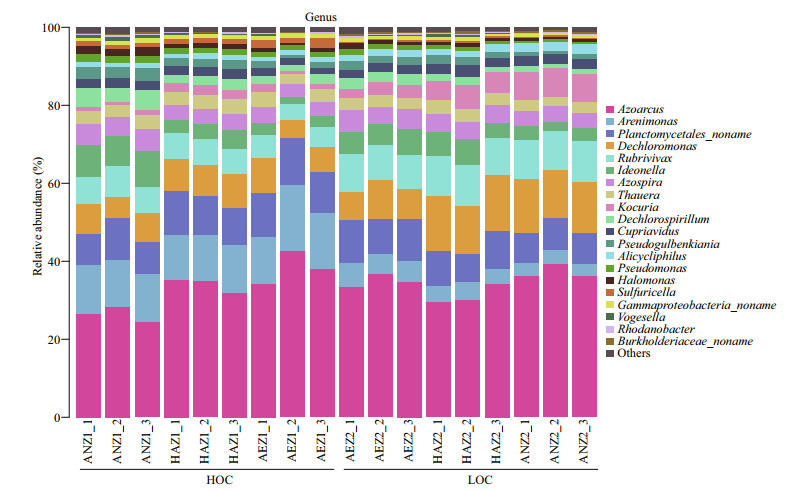
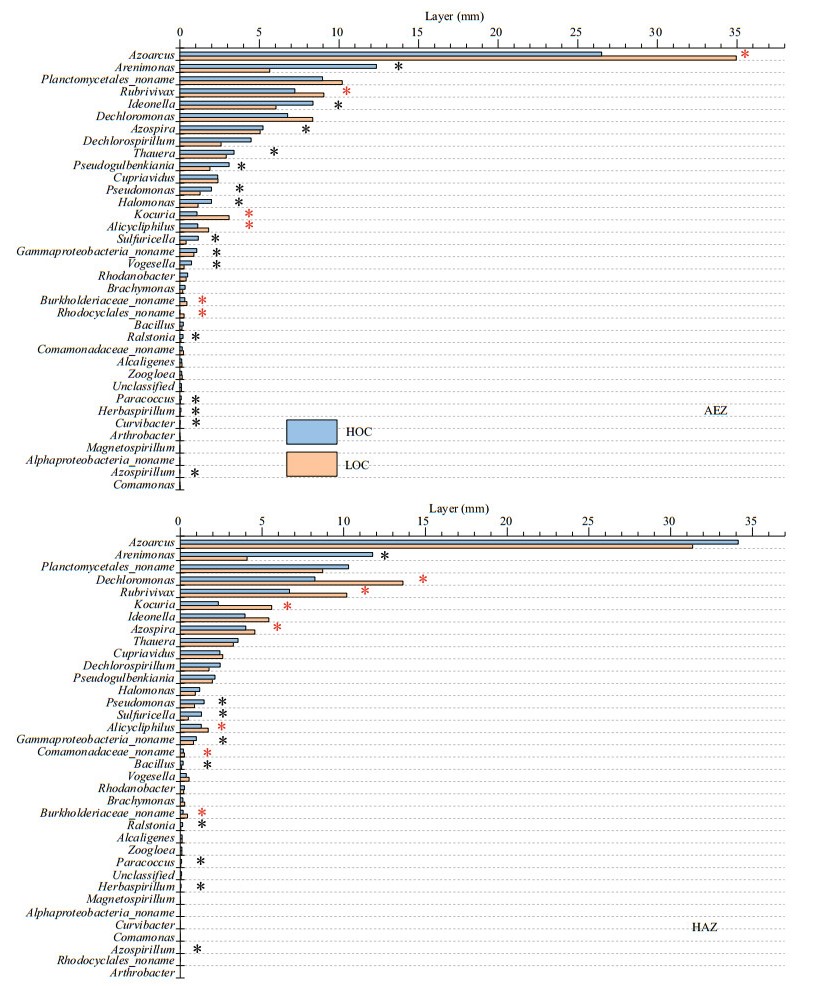
Illumina Miseq-based sequencing identified 36 genera in all the samples, among which only six genera were classified as unknown genera. Azoarcus, Arenimonas, Planctomycetales_noname, Dechloromonas and Rubrivivax were the predominant denitrifier representatives at the surface sediment (Fig. 3). When the bacterial species abundance at the genus level were compared between the high-carbon and low-carbon sediments, there was significant increase in 6 out of 36, 7 out of 36 and 6 out of 36 genera respectively for the aerobic, hypoxic-anoxic and anoxic layers. On the other hand, 14 out of 36, 9 out of 36 and 15 out of 36 genera showed significant decrease in bacterial species abundance respectively for the aerobic, hypoxic-anoxic and anoxic layers. Additionally, 14 out of 36, 20 out of 36, and 15 out of 36 genera had no change in bacterial species abundance respectively for the aerobic, hypoxic-anoxic and anoxic layers. The numbers of bacteria at the genus level were compared. Among them, the dominant genera Azoarcus, Rubrivivax, Kocuria and Alicycliphilus significantly increased, Arenimonas, Ideonella, Dechlorospirillum and Thauera decreased, while Planctomycetales_ noname, Dechloromonas, Azospira and Cupriavidus did not show any significant change at the aerobic layer (0–2 mm). At the hypoxic-anoxic layer (2–4 mm), the predominant genera Dechloromonas, Rubrivivax, Kocuria and Azospira significantly increased, Arenimonas, Pseudomonas and Sulfuricella decreased while Azoarcus, Planctomycetales_noname, Ideonella and Thauera had no observable changes with respect to the consumption of organic carbon. With respect to the consumption of organic carbon, the ascendant genera Dechloromonas, Rubrivivax, Kocuria, and Cupriavidus significantly increased, Alphaproteobacteria_noname, Magnetospirillum, Arenimonas, and Planctomycetales_ noname significantly decreased, while Dechlorospirillum, Azoarcus, Azospira, and Thauera did not show any significant change at the anoxic layer (4–6 mm) (Fig. 4).
|
| Fig.3 Relative abundance of nirS-based representative denitrifier communities in the surface sediment AEZ: aerobic layer; HAZ: hypoxic-anoxic layer; ANZ: anoxic layer; HOC: high organic carbon; LOC: low organic carbon. |
|
| Fig.4 Comparison of the compositions of denitrifier communities of HOC and LOC content within the sediment at three layers (aerobic, AEZ; hypoxic-anoxic, HAZ; and anoxic, ANZ) The relative abundances (percentage) of the microbiota at genera level are presented. Red and black asterisks indicate significant increase and decrease form HOC to LOC, respectively (*: P≤0.05). HOC: high organic carbon; LOC: low organic carbon. |
ANOSIM was used to analyze the differences in the community structure of denitrifiers across the vertical profiles with the consumption of organic carbon in the surface sediment. These comparisons revealed significant differences in denitrifying community structure (R2=0.93, P=0). NMDS also showed significant difference among vertical profiles and between high and low organic carbon sediment surface at the OTU level. On the other hand, the bacterial communities at the same layer and for the same organic carbon content tended to cluster together, with high organic carbon content visibly separated from low organic carbon content, and aerobic (0–2 mm), hypoxic-anoxic (2–4 mm) and anoxic (4–6 mm) clearly parted (Fig. 5).

|
| Fig.5 NMDS showed differences in nirS-based representative denitrifier communities among the three layers (AEZ, HAZ and ANZ) between HOC and LOC AEZ: aerobic layer; HAZ: hypoxic-anoxic layer; ANZ: anoxic layer; HOC: high organic carbon; LOC: low organic carbon. |
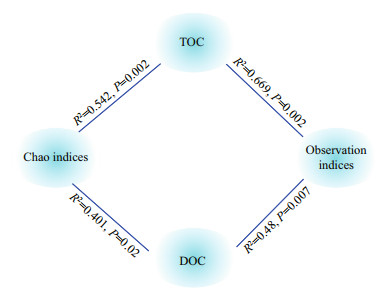
Based on nirS-type Illumina Miseq-based sequencing results, the alpha diversity indices, Observation and Chao, were used together to evaluate the denitrifiers community diversity along the surface sediment under the organic carbon consumption. Two-way ANOVA showed that the Observation indices (P < 0.001) and Chao indices (P < 0.01) differed significantly between the high and low organic carbon content in the surface sediment. With respect to the consumption of organic carbon, the alpha diversity indices increased at the aerobic (0–2 mm) and anoxic layers (4–6 mm) and decreased at hypoxic-anoxic layers (2–4 mm). Under high organic carbon content, the indices showed peak values at the hypoxic-anoxic layers (2–4 mm), and decreased with the increase in the DO levels at the low organic carbon content. The interaction of depth and organic carbon consumption of the Observation (P < 0.001) and Chao (P < 0.01) indices were significantly different (Table 2). Moreover, Pearson's correlation analysis revealed that TOC content and DOC content were also potential nutrients that were significantly correlated with the diversity indices (Fig. 6).
|
|
| Fig.6 Pearson's correlation between organic carbon (TOC and DOC) and the indices of Observation and Chao |
Nitrate loading and contamination of surface waters (e.g., lakes) has become a common environmental and health problem, especially in developing countries. Microbiological processes can remove excess nitrogen in waterbodies. These processes occur mainly at the surface of the sediment, where microbial activities are higher (Saarenheimo et al., 2017). In this study, the consumption of organic carbon is related to the abundance community composition, community structure and diversity of denitrifying bacteria in surface sediments, and there is a significant difference in stratified distribution of denitrifying bacteria under different DO concentrations.
4.1 Effects of organic carbon consumption on denitrifying bacteria in surface sedimentsThe values of different forms (TOC and DOC) of organic carbon affect denitrification in two ways. First, it can provide the substrate needed for the growth of bacteria, which consequently influences the denitrifying bacteria richness. Secondly, the decomposition of organic matter depletes oxygen. Generally, lower oxygen content was required for denitrification. Therefore, high organic concentration will create a favorable environment for denitrification (Knowles, 1982; Lee and Rittmanm, 2003; Wu et al., 2019). In the present study, the abundance of nirS gene decreased significantly with the organic carbon consumption, indicating the potential weakening of denitrification ability because of the continuous utilization of organic carbon of lake surface sediment by bacteria.
Microbial community composition, abundance and diversity vary with the environmental gradients (Bier et al., 2015; Fan et al., 2016), and these changes could be explained by the mechanisms of community adaptability to the variation of ecosystem functioning (Liu et al., 2018b). In this study, 36 representatives of denitrifiers were identified by nirS sequencing. Azoarcus Arenimonas, Planctomycetales_noname, Dechloromonas, and Rubrivivax were the dominant genera within the lake sediment surface. Such result have also been achieved in agricultural soils, activated sludge, landfill leachate and coking wastewater (Thomsen et al., 2007; Remmas et al., 2016; Coyotzi et al., 2017; Li and Lu, 2017). Out of the 36 genera identified through nirS sequencing, six were not identified at the level of genus. This confirms the existence of many potentially novel bacterial populations that need to be explored and identified. Among the identified genera, the dominant Rubrivivax increased significantly with the consumption of organic carbon for the three DO layers at the sediment surface, indicating that the genus could be properly adapted properly to low carbon content. This could be maintaining its high abundance within the lake sediment surface. In biofilm reactors, this genus has also showed a relatively high abundance with degradation of exogenous carbon (Qiu et al., 2017). In the present study, the dominant genera of Arenimonas decreased with respect to the consumption of organic carbon in three DO layers within the surface sediment. This indicates that the genus may be driving denitrification process at high organic carbon content, which consequently affects their growth under the limitation of organic carbon content. This study also showed no significant change in the genera Azoarcus and Ideonella within the hypoxia-anoxic layer (2–4 mm) and anoxic layer (4–6 mm). This revealed that the content of organic carbon had no effect on them within the study conditions. Environmental change is a selective force for the sensitive bacterial individuals, which leads to changes in species and may have cascaded effects on ecosystem functions (Carlisle and Clements, 2005). In the present study, the change of organic carbon concentration in eutrophic lake sediment had selective effect on the species and abundance of denitrifying bacteria.
The influence of organic matter to denitrifier community structure and diversity has often been mentioned in variable ecosystems (Lu et al., 2014; Chen et al., 2018; Si et al., 2018; Xu et al., 2018). However, there are few studies on denitrifying bacterial community structure and diversity in surface sediments with organic matter consumption under different DO concentrations. In this study, the structure of nirS-based denitrifier communities in the surface sediments showed significant difference between high and low organic carbon content in the surface sediment based on NMDS analysis. Chao and Observation indices also showed variation among the different DO layers with the organic carbon consumption. The interspecific competition and coexistence among genera and species of bacteria have been proposed by previous studies (Schmid et al., 2003; Kartal et al., 2007). In the surface sediment, denitrifying bacteria could compete for nutrient with other bacteria and lead to variability in the diversity indices. The significant correlations of DOC and TOC with the diversity indices also hinted as the key factor influencing denitrifier diversity in sediment surface. However, the effect of organic carbon on the diversity of denitrifying bacteria and the source of organic carbon such as submerged plant degradation, algal decomposition, or exogenic organic matter input in eutrophic sediment surface needs to be assessed further.
4.2 Vertical distribution of denitrifying bacteria in different dissolved oxygen layersThe vertical distribution of anaerobic ammoniaoxidizing bacteria under different DO concentrations in lake sediment has been studied and reported (Qin et al., 2018). However, the study on denitrifying bacteria based on DO content within the surface sediments has not been adequately reported. It is generally believed that denitrification can only proceed normally when the DO concentration is kept below 0.5 mg/L in the system (McKenney et al., 2001). In this study, the depth of DO penetration was about 2.0–2.5 mm within the surface sediment, and the oxygen content was below ~0.5 mg/L from a depth of 2.0 mm. The depth of DO penetration recorded here is similar to depths that have been reported in other studies (Laverman et al., 2007; Wang et al., 2014). Based on these findings, we believed that it may be prudent to refer to the zone above a depth of 2 mm as the aerobic layer and below the depth of 2 mm as the hypoxia and anoxic layer based on this study.
The nirS-type denitrifers are more widely distributed in natural environments and is therefore more frequently used as a biomarker of denitrifying bacteria within the sediments (Braker et al., 2001; Bulow et al., 2008). The abundance of nirS denitrifiers changes with water and sediment depth (Kim et al., 2011; Mao et al., 2017). In the present study, the abundance of nirS gene was higher in hypoxia and anoxic layer than in the other layers within the vertical DO profiles, indicating that denitrifers were most abundant in this hypoxia and anoxic layer. Some studies have also shown that the penetration depth of nitrate is greater than that of oxygen based on microelectrode measurements, inferring that the dominant denitrification layer should be below the oxygen layer (Christensen et al., 1989; Sweerts and de Beer, 1989; Nielsen et al., 1990a).
The change in DO concentration can alter the bacterial community structure. When the DO concentration is from 0–0.7 mg/L, then variations of microbial community structures can be realized in a sulfide oxidation and nitrate reduction reactor (Wang et al., 2016). The 2.5 mg/L DO concentration also ensures the optimal bacterial community in a moving bed sequencing batch reactor (Cao et al., 2017). In the present study, the structure of nirS-based denitrifier communities in the surface sediments showed distinctive difference among DO profiles both on high and low organic carbon consumption based on ANOSIM and NMDS analyses. This was consistent with vertical structured patterns of denitrification activity within aquifers (Ben Maamar et al., 2015). This indicated the significant variation of denitrifying community structure formed at aerobic (0.5– 6.9 mg/L), hypoxic and anoxic (0–0.5 mg/L) and anoxic (0 mg/L) DO layers within lake sediment.
5 CONCLUSIONIn this study, specific denitrifier communities existed in different DO layers and the adaptive changes of denitrifier communities were formed with the consumption of organic carbon. The change of organic carbon concentration in eutrophic lake sediment had selective effect on the species and abundance of denitrifying bacteria. In addition, the hypoxic-anoxic layer (2–4 mm) appears to be the main distribution area of denitrifying bacteria in the surface sediments. These findings provide new insights into niche separation based on characteristics of denitrifier communities within the vertical column of lake surface sediment over a few millimeters.
6 DATA AVAILABILITY STATEMENTSequence data that supports the findings of this study have been deposited in NCBI short-read archive under SRA accession PRJNA507510 (https://www.ncbi.nlm.nih.gov/sra/PRJNA507510) and PRJNA5313539 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA531353).
Ben Maamar S, Aquilina L, Quaiser A, Pauwels H, MichonCoudouel S, Vergnaud-Ayraud V, Labasque T, Roques C, Abbott B W, Dufresne A. 2015. Groundwater isolation governs chemistry and microbial community structure along hydrologic flowpaths. Front. Microbiol., 6: 1 457.
DOI:10.3389/fmicb.2015.01457 |
Bier R L, Voss K A, Bernhardt E S. 2015. Bacterial community responses to a gradient of alkaline mountaintop mine drainage in Central Appalachian streams. ISME J., 9(6): 1 378-1 390.
DOI:10.1038/ismej.2014.222 |
Braker G, Ayala-del-Río H L, Devol A H, Fesefeldt A, Tiedje J M. 2001. Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase(nirS) and 16S rRNA genes. Appl. Environ. Microbiol., 67(4): 1 893-1 901.
DOI:10.1128/AEM.67.4.1893-1901.2001 |
Braker G, Fesefeldt A, Witzel K P. 1998. Development of PCR primer systems for amplification of nitrite reductase genes(nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol., 64(10): 3 769-3 775.
DOI:10.1128/AEM.64.10.3769-3775.1998 |
Bulow S E, Francis C A, Jackson G A, Ward B B. 2008. Sediment denitrifier community composition and nirS gene expression investigated with functional gene microarrays. Environ. Microbiol., 10(11): 3 057-3 069.
DOI:10.1111/j.1462-2920.2008.01765.x |
Cao Y F, Zhang C S, Rong H W, Zheng G L, Zhao L M. 2017. The effect of dissolved oxygen concentration (DO) on oxygen diffusion and bacterial community structure in moving bed sequencing batch reactor (MBSBR). Water Res., 108: 86-94.
DOI:10.1016/j.watres.2016.10.063 |
Carlisle D M, Clements W H. 2005. Leaf litter breakdown, microbial respiration and shredder production in metalpolluted streams. Freshw. Biol., 50(2): 380-390.
DOI:10.1111/j.1365-2427.2004.01323.x |
Chen X C, Huang Y Y, Chen G Q, Li P P, Shen Y S, Davis T W. 2018. The secretion of organics by living Microcystis under the dark/anoxic condition and its enhancing effect on nitrate removal. Chemosphere, 196: 280-287.
DOI:10.1016/j.chemosphere.2017.12.197 |
Chen Z G, Wang X J, Yang Y Y, Mirino M W, Yuan Y L. 2016. Partial nitrification and denitrification of mature landfill leachate using a pilot-scale continuous activated sludge process at low dissolved oxygen. Bioresour. Technol., 218: 580-588.
DOI:10.1016/j.biortech.2016.07.008 |
Christensen P B, Nielsen L P, Revsbech N P, Sørensen J. 1989. Microzonation of denitrification activity in stream sediments as studied with a combined oxygen and nitrous oxide microsensor. Appl. Environ. Microbiol., 55(5): 1 234-1 241.
DOI:10.1128/AEM.55.5.1234-1241.1989 |
Coyotzi S, Doxey A C, Clark I D, Lapen D R, Van Cappellen P, Neufeld J D. 2017. Agricultural soil denitrifiers possess extensive nitrite reductase gene diversity. Environ.Microbiol., 19(3): 1 189-1 208.
DOI:10.1111/1462-2920.13643 |
Fan M C, Lin Y B, Huo H B, Liu Y, Zhao L, Wang E T, Chen W M, Wei G H. 2016. Microbial communities in riparian soils of a settling pond for mine drainage treatment. Water Res., 96: 198-207.
DOI:10.1016/j.watres.2016.03.061 |
Francis C A, O'Mullan G D, Cornwell J C, Ward B B. 2013. Transitions in nirS-type denitrifier diversity, community composition, and biogeochemical activity along the Chesapeake Bay estuary. Front. Microbiol., 4: 237.
DOI:10.3389/fmicb.2013.00237 |
Gao J, Hou L J, Zheng Y L, Liu M, Yin G Y, Li X F, Lin X B, Yu C D, Wang R, Jiang X F, Sun X R. 2016. nirS-encoding denitrifier community composition, distribution, and abundance along the coastal wetlands of China. Appl.Microbiol. Biotechnol., 100(19): 8 573-8 582.
DOI:10.1007/s00253-016-7659-5 |
Groffman P M, Butterbach-Bahl K, Fulweiler R W, Gold A J, Morse J L, Stander E K, Tague C, Tonitto C, Vidon P. 2009. Challenges to incorporating spatially and temporally explicit phenomena (hotspots and hot moments) in denitrification models. Biogeochemistry, 93(1-2): 49-77.
DOI:10.1007/s10533-008-9277-5 |
Huang S, Chen C, Yang X, Wu Q, Zhang R. 2011. Distribution of typical denitrifying functional genes and diversity of the nirS-encoding bacterial community related to environmental characteristics of river sediments. Biogeosciences, 8: 3 041-3 051.
DOI:10.5194/bg-8-3041-2011 |
Jenerette G D, Chatterjee A. 2012. Soil metabolic pulses:water, substrate, and biological regulation. Ecology, 93(5): 959-966.
DOI:10.1890/11-1527.1 |
Jia Z M, Liu T, Xia X H, Xia N. 2016. Effect of particle size and composition of suspended sediment on denitrification in river water. Sci. Total Environ., 541: 934-940.
DOI:10.1016/j.scitotenv.2015.10.012 |
Kandeler E, Deiglmayr K, Tscherko D, Bru D, Philippot L. 2006. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol., 72(9): 5 957-5 962.
DOI:10.1128/AEM.00439-06 |
Kartal B, Rattray J, van Niftrik L A, van de Vossenberg J, Schmid M C, Webb R I, Schouten S, Fuerst J A, Damsté J S, Jetten M S M, Strous M. 2007. Candidatus "Anammoxoglobus propionicus" a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol., 30(1): 39-49.
DOI:10.1016/j.syapm.2006.03.004 |
Kim O S, Imhoff J F, Witzel K P, Junier P. 2011. Distribution of denitrifying bacterial communities in the stratified water column and sediment-water interface in two freshwater lakes and the Baltic Sea. Aquat. Ecol., 45(1): 99-112.
DOI:10.1007/s10452-010-9335-7 |
Knowles R. 1982. Denitrification. Microbiol. Rev., 46(1): 43-70.
DOI:10.1128/MMBR.46.1.43-70.1982 |
Laverman A M, Canavan R W, Slomp C P, van Cappellen P. 2007. Potential nitrate removal in a coastal freshwater sediment (Haringvliet Lake, The Netherlands) and response to salinization. Water Res., 41(14): 3 061-3 068.
DOI:10.1016/j.watres.2007.04.002 |
Lee K C, Rittmann B E. 2003. Effects of pH and precipitation on autohydrogenotrophic denitrification using the hollowfiber membrane-biofilm reactor. Water Res., 37(7): 1 551-1 556.
DOI:10.1016/S0043-1354(02)00519-5 |
Li E C, Lu S G. 2017. Denitrification processes and microbial communities in a sequencing batch reactor treating nanofiltration (NF) concentrate from coking wastewater. Water Sci. Technol., 76(11-12): 3 289-3 298.
DOI:10.2166/wst.2017.493 |
Liu J X, Li C, Jing J H, Zhao P Y, Luo Z M, Cao M W, Ma Z Z, Jia T, Chai B F. 2018b. Ecological patterns and adaptability of bacterial communities in alkaline copper mine drainage. Water Res., 133: 99-109.
DOI:10.1016/j.watres.2018.01.014 |
Liu W Z, Yao L, Jiang X L, Guo L D, Cheng X L, Liu G H. 2018a. Sediment denitrification in Yangtze lakes is mainly influenced by environmental conditions but not biological communities. Sci. Total. Environ., 616-617: 978-987.
DOI:10.1016/j.scitotenv.2017.10.221 |
Lu H J, Chandran K, Stensel D. 2014. Microbial ecology of denitrification in biological wastewater treatment. Water Res., 64: 237-254.
DOI:10.1016/j.watres.2014.06.042 |
Mao G Z, Chen L, Yang Y Y, Wu Z, Tong T L, Liu Y, Xie S G. 2017. Vertical profiles of water and sediment denitrifiers in two plateau freshwater lakes. Appl. Microbiol.Biotechnol., 101(8): 3 361-3 370.
DOI:10.1007/s00253-016-8022-6 |
McKenney D J, Drury C F, Wang S W. 2001. Effects of oxygen on denitrification inhibition, repression, and derepression in soil columns. Soil Sci. Soc. Am. J., 65: 126-132.
DOI:10.2136/sssaj2001.651126x |
Nancharaiah Y V, Joshi H M, Hausner M, Venugopalan V P. 2008. Bioaugmentation of aerobic microbial granules with Pseudomonas putida carrying TOL plasmid. Chemosphere, 71(1): 30-35.
DOI:10.1016/j.chemosphere.2007.10.062 |
Nielsen L P, Christensen P B, Revsbech N P, Sørensen J. 1990a. Denitrification and oxygen respiration in biofilms studied with a microsensor for nitrous oxide and oxygen. Microb. Ecol., 19(1): 63-72.
DOI:10.1007/BF02015054 |
Nielsen L P, Christensen P B, Revsbech N P, Sørensen J. 1990b. Denitrification and photosynthesis in stream sediment studied with microsensor and wholecore techniques. Limnol. Oceanogr., 35(5): 1 135-1 144.
DOI:10.4319/lo.1990.35.5.1135 |
Nolan B T. 2001. Relating nitrogen sources and aquifer susceptibility to nitrate in shallow ground waters of the United States. Groundwater, 39(2): 290-299.
DOI:10.1111/j.1745-6584.2001.tb02311.x |
Puckett L J, Tesoriero A J, Dubrovsky N M. 2011. Nitrogen contamination of surficial aquifers-a growing legacy. Environ. Sci. Technol., 45(3): 839-844.
DOI:10.1021/es1038358 |
Qin H Y, Han C, Jin Z W, Wu L, Deng H, Zhu G W, Zhong W H. 2018. Vertical distribution and community composition of anammox bacteria in sediments of a eutrophic shallow lake. J. Appl. Microbiol., 125(1): 121-132.
DOI:10.1111/jam.13758 |
Qiu T L, Xu Y, Gao M, Han M L, Wang X M. 2017. Bacterial community dynamics in a biodenitrification reactor packed with polylactic acid/poly (3-hydroxybutyrate-co-3-hydroxyvalerate) blend as the carbon source and biofilm carrier. J. Biosci. Bioeng., 123(5): 606-612.
DOI:10.1016/j.jbiosc.2016.12.007 |
Remmas N, Melidis P, Katsioupi E, Ntougias S. 2016. Effects of high organic load on amoA and nirS gene diversity of an intermittently aerated and fed membrane bioreactor treating landfill leachate. Bioresour. Technol., 220: 557-565.
DOI:10.1016/j.biortech.2016.09.009 |
Rong N, Shan B Q, Wang C. 2016. Determination of sediment oxygen demand in the Ziya River watershed, China:based on laboratory core incubation and microelectrode measurements. Int. J. Environ. Res. Public Health, 13(2): 232.
DOI:10.3390/ijerph13020232 |
Saarenheimo J, Aalto S L, Rissanen A J, Tiirola M. 2017. Microbial community response on wastewater discharge in boreal lake sediments. Front. Microbiol., 8: 750.
DOI:10.3389/fmicb.2017.00750 |
Santschi P, Höhener P, Benoit G, Buchholtz-ten Brink M. 1990. Chemical processes at the sediment-water interface. Mar. Chem., 30: 269-315.
DOI:10.1016/0304-4203(90)90076-O |
Schmid M, Walsh K, Webb R, Rijpstra W I, van de PasSchoonen K, Verbruggen M J, Hill T, Moffett B, Fuerst J, Schouten S, Sinninghe Damsté J S, Harris J, Shaw P, Jetten M, Strous M. 2003. Candidatus "Scalindua brodae", sp. nov., Candidatus "Scalindua wagneri", sp.nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol., 26(4): 529-538.
DOI:10.1078/072320203770865837 |
Seitzinger S, Harrison J A, Böhlke J K, Bouwman A F, Lowrance R, Peterson B, Tobias C, Van Drecht G. 2006. Denitrification across landscapes and waterscapes:a synthesis. Ecol. Appl., 16(6): 2 064-2 090.
DOI:10.1890/1051-0761(2006)016[2064:DALAWA]2.0.CO;2 |
Si Z H, Song X S, Wang Y H, Cao X, Zhao Y F, Wang B D, Chen Y, Arefe A. 2018. Intensified heterotrophic denitrification in constructed wetlands using four solid carbon sources:denitrification efficiency and bacterial community structure. Bioresour. Technol., 267: 416-425.
DOI:10.1016/j.biortech.2018.07.029 |
Sirivedhin T, Gray K A. 2006. Factors affecting denitrification rates in experimental wetlands:field and laboratory studies. Ecol. Eng., 26(2): 167-181.
DOI:10.1016/j.ecoleng.2005.09.001 |
Srinandan C S, D'souza G, Srivastava N, Nayak B B, Nerurkar A S. 2012. Carbon sources influence the nitrate removal activity, community structure and biofilm architecture. Bioresour. Technol., 117: 292-299.
DOI:10.1016/j.biortech.2012.04.079 |
Sweerts J P R A, de Beer D. 1989. Microelectrode measurements of nitrate gradients in the littoral and profundal sediments of a meso-eutrophic lake (lake Vechten, the Netherlands). Appl. Environ. Microbiol., 55(3): 754-757.
|
Thomsen T R, Kong Y H, Nielsen P H. 2007. Ecophysiology of abundant denitrifying bacteria in activated sludge. FEMS Microbiol. Ecol., 60(3): 370-382.
DOI:10.1111/j.1574-6941.2007.00309.x |
Tian C C, Wang C B, Tian Y Y, Wu X Q, Xiao B D. 2015. Vertical distribution of Fe and Fe(Ⅲ)-reducing bacteria in the sediments of Lake Donghu, China. Can. J. Microbiol., 61(8): 575-583.
DOI:10.1139/cjm-2015-0129 |
Van Rijn J, Tal Y, Schreier H J. 2006. Denitrification in recirculating systems:theory and applications. Aquac.Eng., 34(3): 364-376.
DOI:10.1016/j.aquaeng.2005.04.004 |
Waki M, Yasuda T, Fukumoto Y, Béline F, Magrí A. 2018. Treatment of swine wastewater in continuous activated sludge systems under different dissolved oxygen conditions:reactor operation and evaluation using modelling. Bioresour. Technol., 250: 574-582.
DOI:10.1016/j.biortech.2017.11.078 |
Wang C, Shan B Q, Zhang H, Rong N. 2014. Analyzing sediment dissolved oxygen based on microprofile modeling. Environ. Sci. Pollut. Res. Int., 21(17): 10 320-10 328.
DOI:10.1007/s11356-014-2875-y |
Wang X W, Zhang Y, Zhang T T, Zhou J T. 2016. Effect of dissolved oxygen on elemental sulfur generation in sulfide and nitrate removal process:characterization, pathway, and microbial community analysis. Appl. Microbiol.Biotechnol., 100(6): 2 895-2 905.
DOI:10.1007/s00253-015-7146-4 |
Wen X, Gong B Z, Zhou J, He Q, Qing X X. 2017. Efficient simultaneous partial nitrification, anammox and denitrification (SNAD) system equipped with a real-time dissolved oxygen (DO) intelligent control system and microbial community shifts of different substrate concentrations. Water Res., 119: 201-211.
DOI:10.1016/j.watres.2017.04.052 |
Wu S F, Wu Z, Liang Z Y, Liu Y, Wang Y L. 2019. Denitrification and the controlling factors in Yunnan Plateau Lakes(China):exploring the role of enhanced internal nitrogen cycling by algal blooms. J. Environ. Sci., 76: 349-358.
DOI:10.1016/j.jes.2018.05.028 |
Wu X, Liu G, Butterbach-Bahl K, Fu B, Zheng X, Brüggemann N. 2013. Effects of land cover and soil properties on denitrification potential in soils of two semi-arid grasslands in Inner Mongolia, China. J. Arid Environ., 92: 98-101.
DOI:10.1016/j.jaridenv.2013.02.003 |
Xu N, Tan G C, Wang H Y, Gai X P. 2016. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil. Biol., 74: 1-8.
DOI:10.1016/j.ejsobi.2016.02.004 |
Xu Z S, Dai X H, Chai X L. 2018. Effect of different carbon sources on denitrification performance, microbial community structure and denitrification genes. Sci. Total Environ., 634: 195-204.
DOI:10.1016/j.scitotenv.2018.03.348 |
Yang J K, Cheng Z B, Li J, Miao L H. 2013. Community composition of nirS-type denitrifier in a shallow eutrophic lake. Microb. Ecol., 66(4): 796-805.
DOI:10.1007/s00248-013-0265-5 |
Zou Y, Xu X C, Wang X J, Yang F L, Zhang S S. 2018. Achieving efficient nitrogen removal and nutrient recovery from wastewater in a combining simultaneous partial nitrification, anammox and denitrification (SNAD)process with a photobioreactor (PBR) for biomass production and generated dissolved oxygen (DO)recycling. Bioresour. Technol., 268: 539-548.
DOI:10.1016/j.biortech.2018.08.015 |
Zumft W G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev., 61(4): 533-616.
DOI:10.1016/j.ccr.2004.08.030 |
 2020, Vol. 38
2020, Vol. 38