Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CHEN Na, LI Li, LI Chenghua, LIN Zhihua, MENG Jie, LIU Sheng, SONG Kai, BAO Yongbo
- bHLH genes polymorphisms and their association with growth traits in the Pacific oyster Crassostrea gigas
- Journal of Oceanology and Limnology, 38(3): 862-868
- http://dx.doi.org/10.1007/s00343-019-9070-4
Article History
- Received Mar. 16, 2019
- accepted in principle Jul. 2, 2019
- accepted for publication Aug. 19, 2019
2 School of Marine Sciences, Ningbo University, Ningbo 315211, China;
3 Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China
The Pacific oyster Crassostrea gigas is an economically and ecologically important mollusk extensive distribution in many countries, such as China and Japan (She et al., 2015; Qi et al., 2017) and it has one of the highest levels of genomic DNA variation (Zhang et al., 2012). With the rapid development of marker-assisted selection (MAS), genetic improvement of many aquaculture species have been achieved (Lo Presti et al., 2010), which greatly improve the efficiency and accuracy of selection in breeding schemes. Single nucleotide polymorphisms (SNPs) are highly abundant and a co-dominant mode of inheritance with ease of high-throughput detection (Qi et al., 2017). This has resulted in widespread use of SNPs in MAS (Morin et al., 2004; Park et al., 2009; Fournier-Level et al., 2011; Gaut, 2012). Therefore, identification of genetic markers associated with growth traits could accelerate C. gigas breeding programs (Cong et al., 2014).
Only a few genes that are significantly associated with growth are detected, because of insufficient biological pathways and genetic knowledge involved in bivalve growth regulation. For example, gene TGF-beta Ⅰ (Guo et al., 2012), an insulin-related peptide, is significantly associated with growth traits of C. gigas, such as shell width, soft mass, and body mass (Cong et al., 2014). The amylase gene of the polymorphism was associated with growth rate in C. gigas (Prudence et al., 2006; Huvet et al., 2008). Furthermore, there are many studies showing that some bHLH genes are associated with growth traits. For example, Lai et al. (2013) found two new mutations in the bHLHe40 gene, and they were significantly correlated with growth traits in cattle. Xue et al. (2011) found a novel SNP in the MyoG gene, and it was significantly correlated with body length in native Chinese cattle breeds. The family of transcription factors bHLH plays an important regulatory role in the process of biological growth and development (Atchley and Fitch, 1997). These genes are involved in regulation of neuronal development, cellular differentiation, growth, hematopoiesis, sex determination, bowel development, and apoptosis (Atchley and Fitch, 1997; Ledent and Vervoort, 2001). Meanwhile, many studies have shown that the MyoD1 gene is associated with growth traits in chickens, pigs, ducks, and other animals (Liu et al., 2008; Wu et al., 2012).
Here we analyzed the bHLH family of genes related to growth traits in oysters, from which we screened a set of candidate genes for association analysis using re-sequencing in a wild population. We then validated the results in an independent population using the SNPshot method. Our research aims to identify the bHLH genes and mutations associated with growth traits, and provide genetic tools for MAS of oysters.
2 MATERIAL AND METHOD 2.1 Animals, traits and DNA extractionA total of 417 wild C.gigas from Qingdao (QD1), China, were collected for discovery SNP for preliminary association analysis. Moreover, 288 wild C. gigas were randomly sampled from Qingdao (QD2). Shell height, length, and width were measured using an electronic Vernier caliper (0.01 mm accuracy). Body mass and soft-tissue mass were weighed with an electronic balance (0.1 g accuracy).
Genomic DNA was obtained from adductor muscles of QD1 and QD2 using a genomic DNA isolation kit (Non-centrifugal columnar, Tiangen, Beijing, China). The extracted DNA was examined by 1% agarose gel electrophoresis, and the DNA purity and concentration was quantitatively determined using A NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). When the concentration is ≥30 ng/μL, the purity level (optical density [OD]: OD260/OD280) is 1.7-2.0, which can then be used. The prepared DNA samples were diluted to 10-30 ng/μL with DEPC water and then stored at -80℃.
2.2 Re-sequencingAn amount of 5 μg DNA of each individual was sheared into fragments of 200-800 bp using the Covaris system (Life Technologies, Carlsbad, CA). DNA fragments were processed according to the Illumina DNA sample preparation protocol. Fragments were end-repaired, A-tailed, ligated to paired-end adaptors, and amplified by PCR to 400- 500 bp inserts for library construction (Qi et al., 2017). Sequencing was performed to generate 100-bp paired-end reads on the HiSeq 2000 platform (Illumina), according to the manufacturer standard protocols.
2.3 Sequence alignment and genotype callingFiltered sequence from all individuals were aligned to the Pacific oyster reference genome (GenBank accession number GCA_000297895.1) using the BWA (Burrows-Wheeler Aligner) software and set up parameters 'mem -M -t 10 -T 20' (Li and Durbin, 2009). Picard-tools (version 1.117) were used to sort and index the bam alignment files. The insertions and deletions (INDELs) in the bam files realignment using the Genome Analysis Toolkit (GATK) (McKenna et al., 2010) module RealignerTargetCreator and IndelRealigner and base quality score recalibration was performed using BaseRecalibrator. Population variant calling was processed by HaplotypeCaller module from GATK (Li et al., 2018). Candidate SNPs were filtered in multiple steps using several criteria to eliminate possible false-positives, and to distribute SNPs relatively evenly across the bHLH family genes. For each oyster, if a SNP had low genotype quality (GQ < 20), or low excessive read coverage (DP < 0 or DP > 100), the SNP genotype call of the individual was considered to be missing or invalid (Qi et al., 2017).
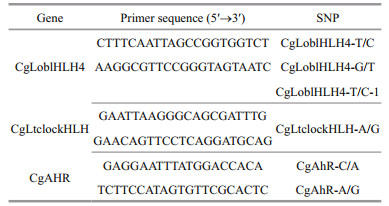
2.4 Validation of candidate SNPsScreened six SNPs were genotyped using an ABI (Applied Biosystems, Foster City, CA, USA) SNPshot method, according to the manufacturer manual. PCR primers (Table 1) were designed according to candidate genes. The DNA sample was obtained from adductor muscles of QD2. The amplification parameters were 95℃ denaturation for 5 min, 94℃ for 30 s, 50-55℃ for 30 s, 72℃ for 45 s for 35 cycles, plus a 72℃ extension step of 10 min for both SNPs (Guo et al., 2015). All SNPs reported in this manuscript had a genotyping success rate > 98% and accuracy > 99%, as judged by random re-genotyping of 20% of the samples in the cohort.
The values of oyster related traits were analyzed by SPSS version 16.0, including the number, mean, maximum, minimum, standard deviation, and coefficient of variation of each trait. SPSS version 16.0 was used to conduct the descriptive statistical analysis of the trait. We also performed a correlation analysis to determine whether SNPs were correlated with shell height, shell length, shell width, body mass, or soft-tissue mass. Growth traits between different genotypes were compared using ANOVA, using SPSS version 16.0.
3 RESULT 3.1 Phenotypic statisticsFive growth index parameters were measured: shell height, shell width, shell length, soft-tissue mass, and body mass (Supplementary Tables S1 & S2); this data were normally distributed (P > 0.05) in QD1 and QD2 wild oysters (Supplementary Figs.S1 & S2).
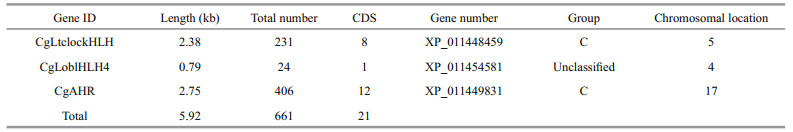
3.2 Re-sequencing and selection of SNPsFor SNP discovery, we re-sequenced 417 wild oysters with coverage of > 20 fold for each individual. After aligning the reads to 88 reference sequences of oyster bHLH through BWA, we identified 4 096 high quality SNPs using Samtools and GATK software. Then, we selected 661 SNPs in candidate genes from the total of bHLH genes (Supplementary Table S3). We identified two genes from the C group of CgAhR and CgLtclockHLH, and one novel CgLoblHLH4 gene. By aligning the reads of the 5.92 kb (Table 2) candidate gene sequence, we identified 62 high quality SNPs from a total of 661 SNPs associated with growth traits (P < 0.05).
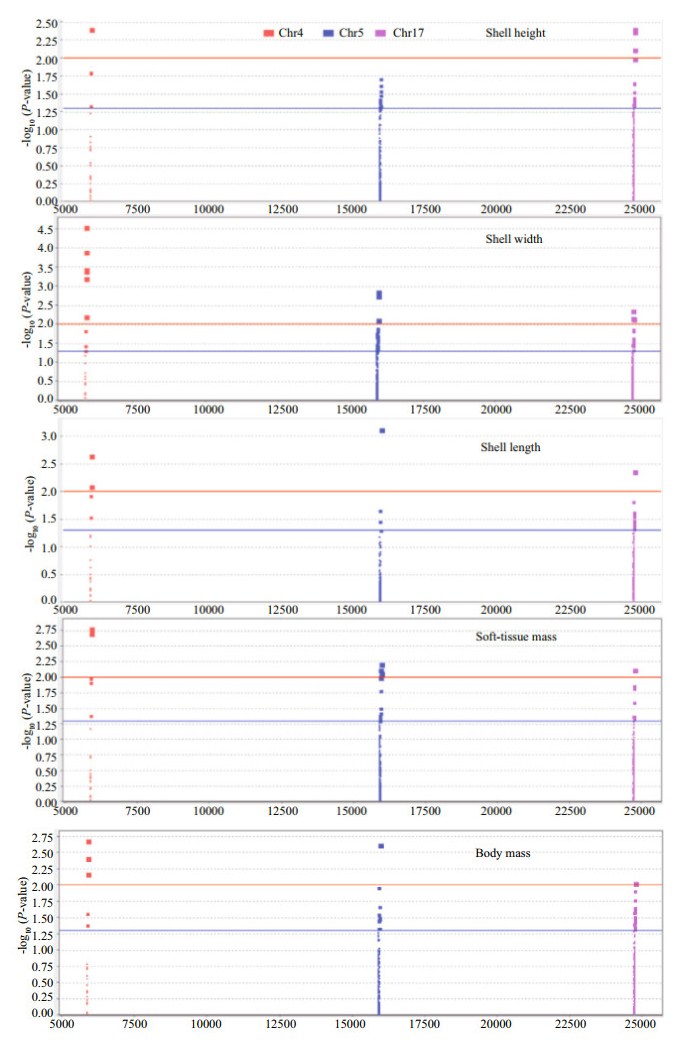
SNPs associated with shell height, shell width, shell length, soft-tissue mass, and body mass that were detected in candidate genes are shown on Manhattan plots (Fig. 1). CgLoblHLH4 in chromosome 4 had 12 high quality SNPs associated with growth traits. CgLtclockHLH in chromosome 5 had 21 high quality SNPs associated with growth traits. CgAhR of chromosome 17 had 29 high quality SNPs associated with growth traits. The horizontal blue dashed line is the threshold; SNPs above this line are significantly associated with growth traits (P < 0.05). The horizontal red dashed line is the threshold; SNPs above this line are very significantly associated with growth traits (P < 0.01). A polymorphism can be associated with several growth traits, so there are repeated points.
|
| Fig.1 The Manhattan plot of SNPs associated with growth traits The x-axis presents genomic coordinates along chromosomes 4, 5 and 17. The y-axis presents a negative logarithm of P-values. The horizontal blue dashed line is the threshold, SNPs above this line are significantly associated with growth traits (P < 0.05). The horizontal red dashed line is the threshold, SNPs above this line are extremely significant associated with growth traits (P < 0.01). CgLoblHLH4 in chromosome4 (Chr4), CgLtclockHLH in chromosome 5 (Chr5), CgAhR in chromosome 17 (Chr17). |
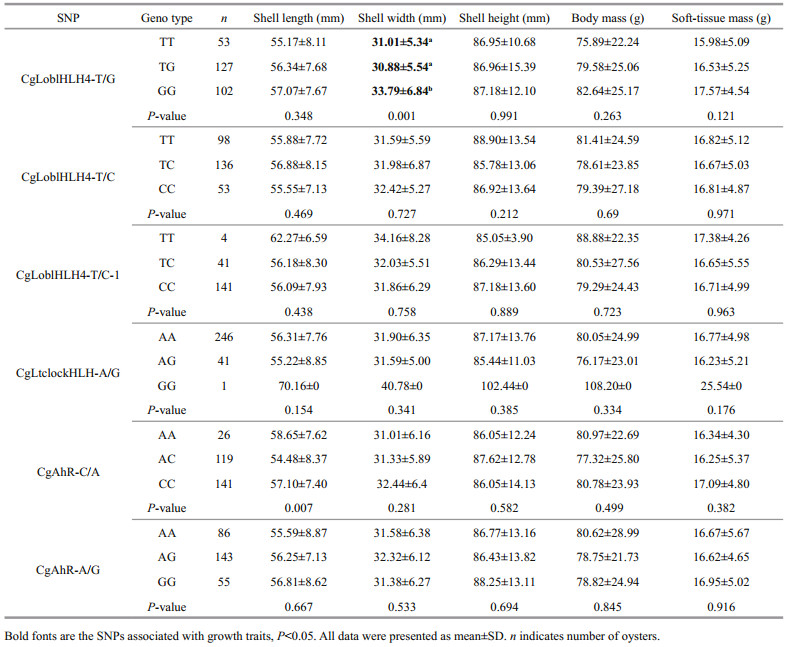
At the second stage of candidate gene association analysis, using the six SNPs genotyped by the SNPshot method (Table 3), we identified one SNP (CgLoblHLH4-T/G) associated with length (P < 0.05). CgLoblHLH4-T/G was located in the exon of CgLoblHLH4. It was a non-synonymous mutation that changed the amino acid composition of Leu to Phe. In QD1 and QD2, shell widths of oysters with GG allele were wider than that of TT and TG (P < 0.05). The remaining 5 SNPs were not significantly associated with growth traits in this oyster population. The SNP CgAhR-C/A genotype AA and CC are both significantly wider than heterozygote AC in shell length is not analysis in our study.
By aligning the reads of the 5.92 kb reference sequence, a SNP calling was made per each 8.93 bp on average, which was sufficient for correlation analysis. It is well known genetic variation is the key to long-term improvement of cultured strains (Zhong et al., 2013; Wang et al., 2014). Therefore, the discovery and effective use of genetic variation is essential for the sustainable development of aquaculture (Guo, 2009). In this study, 661 SNPs were identified in the coding study region of the candidate genes, which is consistent with previous study that the oysters have one of the highest levels of genomic polymorphism in animals (Zhang et al., 2012).
CgLoblHLH4-T/G was located in the exon of the CgLoblHLH4 gene. In QD1 and QD2, individuals with GG shell width were higher than those with TT and TG shell width, and have non-synonymous mutations, resulting in a change of an amino acid in the coding region from Phe to Leu, the Leu allele appears to be associated with an increase in shell width. It is speculated that the encoded amino acid is altered by changing the protein structure and biological activity, lead to oysters have better growth traits. The CgLoblHLH4 gene is a novel gene specific for lophotrochozoan. Bao et al. (2017) found that the CgLoblHLH gene family is duplicated in C. gigas, yielding four genes. The expression of these genes in early development, as determined by transcriptome data analysis, is dynamic, it appears to change successively through CgLoblHLH4 in oocytes and cleavage stage embryo, to CgLoblHLH2 in gastrula stages, and to CgLoblHLH1 by the trochophore stage (Bao et al., 2017). These findings have been confirmed in our current research (unpublished). Therefore, it is reasonable to speculate that the CgLoblHLH4 gene plays an important role in early development stage of C. gigas, and determines the shell width index of C. gigas. However, there are no reports on the regulatory mechanism of CgLoblHLH4 gene for growth regulation in C. gigas, which need for further study.
5 CONCLUSIONWe detected SNPs in 88 candidate genes from the bHLH family and their association with growth traits. Firstly, 661 SNPs were called by re-sequencing candidate genes in QD1. In the second stage, six SNPs were genotyped by SNPshot method, and finally CgLoblHLH4-T/G in the CgLoblHLH4 gene was found to be related to shell width. In addition, the shell of C.gigas with the GG genotype was wider than that of oysters with TT and TG genotypes. Oysters with GG genotype have growth advantages for breeding. These findings not only provide an insight into the genetic basis of C. gigas quality traits, but also provide molecular markers for MAS of oysters.
6 DATA AVAILABILITY STATEMENTThe whole-genome re-sequencing and transcriptome datasets are deposited in the Sequence Read Archive (SRA) database under the accession number PRJNA394055.
Atchley W R, Fitch W M. 1997. A natural classification of the basic helix-loop-helix class of transcription factors. Proceedings of the National Academy of Sciences of the United States of America, 94(10): 5 172-5 176.
DOI:10.1073/pnas.94.10.5172 |
Bao Y B, Xu F, Shimeld S M. 2017. Phylogenetics of lophotrochozoan bHLH genes and the evolution of lineage-specific gene duplicates. Genome Biology and Evolution, 9(4): 869-886.
DOI:10.1093/gbe/evx047 |
Cong R H, Kong L F, Yu H, Li Q. 2014. Association between polymorphism in the insulin receptor -related receptor gene and growth traits in the Pacific oyster Crassostrea gigas. Biochemical Systematics and Ecology, 54: 144-149.
DOI:10.1016/j.bse.2014.02.003 |
Fournier-Level A, Korte A, Cooper M D, Nordborg M, Schmitt J, Wilczek A M. 2011. A map of local adaptation in Arabidopsis thaliana. Science, 334(6052): 86-89.
DOI:10.1126/science.1209271 |
Gaut B. 2012. Arabidopsis thaliana as a model for the genetics of local adaptation. Nature Genetics, 44(2): 115-116.
DOI:10.1038/ng.1079 |
Guo H H, Bao Z M, Li J Q, Lian S S, Wnag S, He Y, Fu X T, Zhang L L, Hu X L. 2012. Molecular characterization of TGF-β type Ⅰ receptor gene (Tgfbr1) in Chlamys farreri, and the association of allelic variants with growth traits. PLoS One, 7(11): e51005.
DOI:10.1371/journal.pone.0051005 |
Guo S X, Hu Y H, Ding Y S, Liu J M, Zhang M, Ma R L, Guo H, Wang K, He J, Yan Y Z, Rui D S, Sun F, Mu L T, Niu Q, Zhang J Y, Li S G. 2015. Association between eight functional polymorphisms and haplotypes in the cholesterol ester transfer protein (CETP) gene and dyslipidemia in national minority adults in the far west region of China. International Journal of Environmental Research and Public Health, 12(12): 15 979-15 992.
DOI:10.3390/ijerph121215036 |
Guo X M. 2009. Use and exchange of genetic resources in molluscan aquaculture. Reviews in Aquaculture, 1(3-4): 251-259.
DOI:10.1111/j.1753-5131.2009.01014.x |
Huvet A, Jeffroy F, Fabioux C, Daniel J Y, Quillien V, Van Wormhoudt A, Moal J, Samain J F, Boudry P, Pouvreau S. 2008. Association among growth, food consumptionrelated traits and amylase gene polymorphism in the Pacific oyster Crassostrea gigas. Animal Genetics, 39(6): 662-665.
DOI:10.1111/j.1365-2052.2008.01776.x |
Lai X S, Zhang C G, Wang J, Wang C, Lan X Y, Lei C Z, Chen H. 2013. Developmental expression patterns and association study with growth traits of bovine Bhlhe40 gene. Molecular Biology, 47(5): 674-680.
DOI:10.1134/S0026893313050105 |
Ledent V, Vervoort M. 2001. The basic helix-loop-helix protein family:comparative genomics and phylogenetic analysis. Genome Research, 11(5): 754-770.
DOI:10.1101/gr.177001 |
Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14): 1 754-1 760.
DOI:10.1093/bioinformatics/btp324 |
Li L, Li A, Song K, Meng J, Guo X M, Li S M, Li C Y, De Wit P, Que H Y, Wu F C, Wang W, Qi H G, Xu F, Cong R H, Huang B Y, Li Y X, Wang T, Tanh X Y, Liu S, Li B S, Shi R H, Liu Y L, Bu C, Zhang C, He W M, Zhao S C, Li H J, Zhang S D, Zhang L L, Zhang G F. 2018. Divergence and plasticity shape adaptive potential of the Pacific oyster. Nature Ecology & Evolution, 2(11): 1 751-1 760.
|
Liu M, Peng J, Xu D Q, Zheng R, Li F E, Li J L, Zuo B, Lei M G, Xiong Y Z, Deng C Y, Jiang S W. 2008. Association of MYF5 and MYOD1 gene polymorphisms and meat quality traits in large white×meishan F2 pig populations. Biochemical Genetics, 46(11-12): 720-732.
DOI:10.1007/s10528-008-9187-1 |
Lo Presti R, Lisa C, Di Stasio L. 2010. Molecular genetics in aquaculture. Italian Journal of Animal Science, 8(3): 299-313.
|
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo M A. 2010. The genome analysis toolkit:a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20(9): 1 297-1 303.
DOI:10.1101/gr.107524.110 |
Morin P A, Luikart G, Wayne R K, the SNP Workshop Group. 2004. SNPs in ecology, evolution and conservation. Trends in Ecology & Evolution, 19(4): 208-216.
|
Park Y J, Lee J K, Kim N S. 2009. Simple sequence repeat polymorphisms (SSRPs) for evaluation of molecular diversity and germplasm classification of minor crops. Molecules, 14(11): 4 546-4 569.
DOI:10.3390/molecules14114546 |
Prudence M, Moal J, Boudry P, Daniel J Y, Quéré C, Jeffroy F, Mingant C, Ropert M, Bédier E, Van Wormhoudt A, Samain J F, Huvet A. 2006. An amylase gene polymorphism is associated with growth differences in the Pacific cupped oyster Crassostrea gigas. Animal Genetics, 37(4): 348-351.
DOI:10.1111/j.1365-2052.2006.01481.x |
Qi H G, Song K, Li C Y, Wang W, Li B S, Li L, Zhang G F. 2017. Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas). PLoS One, 12(3): e0174007.
DOI:10.1371/journal.pone.0174007 |
She Z C, Li L, Qi H G, Song K, Que H Y, Zhang G F. 2015. Candidate gene polymorphisms and their association with glycogen content in the pacific oyster Crassostrea gigas. PLoS One, 10(5): e0124401.
DOI:10.1371/journal.pone.0124401 |
Wang J F, Qi H G, Li L, Que H Y, Wang D, Zhang G F. 2014. Discovery and validation of genic single nucleotide polymorphisms in the Pacific oyster Crassostrea gigas. Molecular Ecology Resources, 15(1): 123-135.
|
Wu Y, Pi J S, Pan A L, Pu Y J, Du J P, Shen J, Liang Z H, Zhang J R. 2012. An SNP in the MyoD1 gene intron 2 associated with growth and carcass traits in three duck populations. Biochemical Genetics, 50(11-12): 898-907.
DOI:10.1007/s10528-012-9530-4 |
Xue M, Zan L S, Gao L, Wang H B. 2011. A novel polymorphism of the myogenin gene is associated with body measurement traits in native Chinese breeds. Genetics & Molecular Research, 10(4): 2 721-2 728.
|
Zhang G F, Fang X D, Guo X M, Li L, Luo R B, Xu F, Yang P C, Zhang L L, Wang X T, Qi H G, Xiong Z Q, Que H Y, Xie Y L, Holland P W H, Paps J, Zhu Y P, Wu F C, Chen Y X, Wang J F, Peng C F, Meng J, Yang L, Liu J, Wen B, Zhang N, Huang Z Y, Zhu Q H, Feng Y, Mount A, Hedgecock D, Xu Z, Liu Y J, Domazet-Lošo T, Du Y S, Sun X Q, Zhang S D, Liu B H, Cheng P Z, Jiang X T, Li J, Fan D D, Wang W, Fu W J, Wang T, Wang B, Zhang J B, Peng Z Y, Li Y, Li N, Wang J P, Chen M S, He Y, Tan F J, Song X R, Zheng Q M, Huang R L, Yang H L, Du X D, Chen L, Yang M, Gaffney P M, Wang S, Luo L H, She Z C, Ming Y, Huang W, Zhang S, Huang B Y, Zhang Y, Qu T, Ni P X, Miao G Y, Wang J Y, Wang Q, Steinberg C E W, Wang H Y, Li N, Qian L M, Zhang G J, Li Y R, Yang H M, Liu X, Wang J, Yin Y, Wang J. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature, 490(7418): 49-54.
DOI:10.1038/nature11413 |
Zhong X X, Li Q, Yu H, Kong L F. 2013. Development and Validation of single-nucleotide polymorphism markers in the pacific oyster, Crassostrea gigas, using high-resolution melting analysis. Journal of the World Aquaculture Society, 44(3): 455-465.
DOI:10.1111/jwas.12044 |
 2020, Vol. 38
2020, Vol. 38