Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Xiuliang, YAO Jianting, ZHANG Jie, DUAN Delin
- Status of genetic studies and breeding of Saccharina japonica in China
- Journal of Oceanology and Limnology, 38(4): 1064-1079
- http://dx.doi.org/10.1007/s00343-020-0070-1
Article History
- Received Jan. 31, 2020
- accepted in principle Mar. 13, 2020
- accepted for publication Apr. 16, 2020
2 Lab for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266071, China
Saccharina japonica is regarded as one of the most important, cultivated brown seaweeds in the world. In China, its large-scale industrialization started from the late 1950s. The current annual, global production is approximately 8 million tons (wet weight) with an approximate value of 4 millions US dollars. In the past, strain selection and breeding, classical quantitative genetic studies and limited cytological studies were summarized to S. japonica by Patwary and van der Meer (1992). At the beginning of the 21st century, based on the advancement of sequencing techniques (Shendure and Ji, 2008), genomics, comparative transcriptomics, proteomics, and QTL mapping were applied to study S. japonica due to its economic value (Hafting et al., 2015). In addition, new varieties of S. japonica have been developed, which provided enhanced characteristics of higher yields or temperature resistance, etc. This review presents the status of genetic and breeding research as applied to S. japonica in China, and the key future prospects in this filed are outlined.
2 CULTIVATION OF S.JAPONICA IN CHINASaccharina japonica is naturally distributed around the sub-littoral areas of the Northwestern Pacific, such as far-eastern Russia, Kamchatka Island, Hokkaido, the Kurile islands, and northern coastal area of Korean in the Japan Sea (Zhang et al., 2015c, 2019b). Previously it was named as Laminaria japonica (Lane et al., 2006). Lane et al. (2006) renamed L. japonica as S.japonica, by restoring it to the genus Saccharina, according to molecular phylogenies using multi-marker systems (Yoon et al., 2001; Lane et al., 2006; Bartsch et al., 2008).
In China, S. japonica is regarded as an alien species. Early in 1927, it was discovered attached to rocks on the coast of Dalian. Later, a Japanese technician, Mr. Yoshiro Otuki conducted the pioneer research on kelp cultivation trials using this species in Yantai in early 1943. However, his initial efforts failed (Fang and Zhang, 1982). Since then, Chinese phycologists and technicians have tackled two major bottlenecks in the cultivation of S. japonica: firstly, the ability to culture sporelings during summer with natural light and cool seawater in the nursery greenhouse, and secondly, the need for long-lines, or floating raft culture systems in the open sea. Oncethese issues were addressed, the industrial cultivation of S.japonica was eventually implemented (Tseng et al., 1955, 1957, 1962; Tseng, 1981; Su et al., 2017) (Fig. 1).

|
| Fig.1 S. japonica life history and cultivation practice in China (rewritten from Duan et al., 2015) |
With the mix of fertile male and female gametophytes, it was regarded as a new way for seedling nursery (Pang and Wu, 1996; Li et al., 1999; Zhang et al., 2008a). If under the suitable cultural conditions, the mixed female and male gametophytes could be induced to fertilize and yield into young sporophytes after 20 days. Although this method was comparatively efficient of time and cost, only limited company adopt this method for cultivars yielding on small scale in China, because of the weak adherence to the substratum to the young seedling at the initial stage, and the seedling lost in the production.
Presently, S. japonica commercialization was mainly conducted in Asia countries, especially in Korea, North Korea, Japan, and China. In 2016, the total wet weight production for this species in China was about 7.3 million tons, valued as 3.89 millions US dollars (FAO, 2018); still there are small scale production in the far east of Russia (mainly in Primorie) (Selivanova et al., 2006).
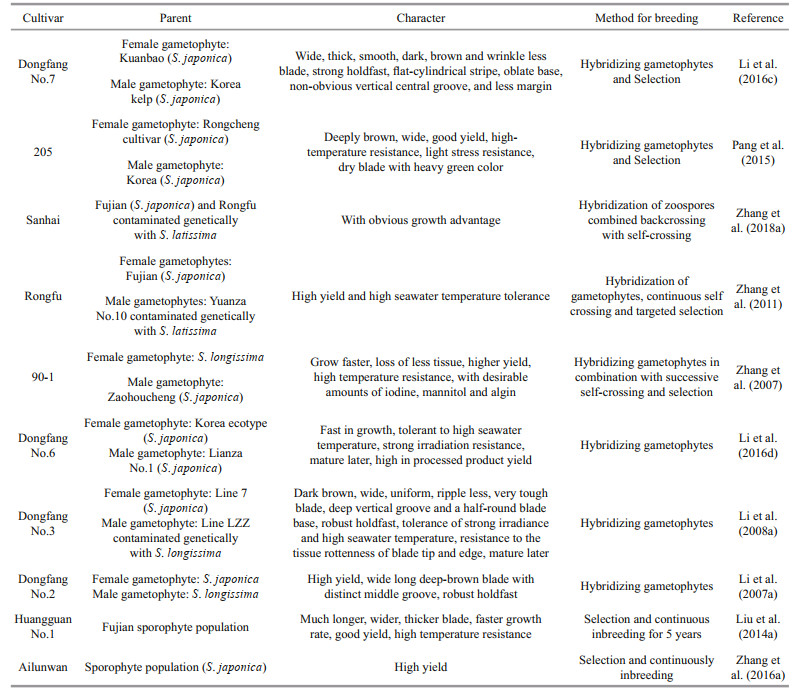
3 SELECTION AND BREEDING OF NEW VARIATIES FOR S. JAPONICAAt the present time, the breeding for new kelp cultivars was mainly performed in China, Korea, and Japan. Hwang et al. (2019) reviewed the breeding progress in the eastern Asian countries mainly for the five species, S. japonica, Pyropia spp., Undaria spp., Cladosiphon okamuranus, and Nemacystus decipiens. To S. japonica, the first cultivar "Haiqing No.1" was bred in 1962, which characterized of faster growth and late sorus matures and resistant to higher illumination (Fang et al., 1963). Recently, about 10 new varieties were selected or hybridized (Table 1), and were applied in the production in China.
For the breeding strategies, one way is directional selection during the continuously inbreeding (Fang et al., 1965), another way is hybridization of the gametophytes according to genetic distance, "Danhai No.1" and "Danza No.10" are the first varieties hybridized (Fang et al., 1983, 1985). With the integration of DNA markers techniques, the molecular assistance in selection and breeding were applied in the kelp breeding, such as parental analysis to Dongfang No.2 (Shi et al., 2008), prediction of heterosis (Li et al., 2008a) and genetic variation to the new cultivars (Li et al., 2012; Zhang et al., 2016a, 2018a). Furthermore, it was adopted for the identification of genotype for the phenotype (Liu et al., 2011; Wang et al., 2018), and we believed that this techniques will play more important role in the kelp selection and breeding.
4 DNA MARKERS AND THEIR APPLICATIONS IN GENETIC ANALYSISUsually, the DNA markers are classified into three categories, hybridization-based, PCR-based and sequence-based (Table 2.1 in Xu, 2010). Previously, without knowing the genomic data of S. japonica, RAPD, AFLP, ISSR and SCAR were adopted for many genetic analysis, such as analyzing different morphological sub-species in Japan (Yotsukura et al., 2001), identifying different gametophytic germplasm varieties (Wang et al., 2004), discriminating female gametophytes (Liu et al., 2009b; Gu et al., 2014; Zhang et al., 2018c), and verifying the parent of one new developed hybrid variety (Shi et al., 2008).
Codominant SSR markers was preferred for population genetic analysis and genetic linkage map construction due to its highly polymorphism. So far, there are three resolutions for developing SSR marker for S. japonica. One way was developing SSR markers directly from SSR-enriched genomic library (Billot et al., 1998; Shi et al., 2007; Zhang et al., 2015a). Another way was developing microsatellites DNA markers through EST databases mining, and generated markers for genetic diversity analysis (Liu et al., 2010b; Wang et al., 2011). Liu et al. (2012b) ever developed 13 SSRs to S.japonica from the 2 668 non-redundant EST sequences of L. digitata. The last method was developing SSRs via the genomic and transcriptomic data (Zhang et al., 2014, 2016b, 2018b, 2019a; Li et al., 2016a; Peng et al., 2016). Li et al. (2016a) identified 181 595 SSR loci using the MISA program, and applied the marker to 24 S. japonica individuals analysis.
Recently, huge SNP markers were developed for genetic linkage map construction and QTL mapping with the next generation sequencing platform (Zhang et al., 2015f; Wang et al., 2018). Generally, it is expected that the high throughout available SNP markers, will be the first candidate for S. japonica genetics analysis in the future.
5 CYTOGENETICS STUDYWith the improvements of squash technique to chromosomes analysis, the chromosome number was characterized in Saccharina related species (Lewis, 1996). Its haploid number estimated were from 22 (Yabu, 1973; Tai and Fang, 1977), 28–35 (Nakahara, 1984), 16–24 (Lewis et al., 1993) and 32 (Yabu and Yasui, 1991) respectively. Zhou et al. (2004) reported the chromosome numbers of haploid male gametophyte was 31. Liu et al. (2012d) either observed the haploid male and female gametophytes was 31, and 62 for diploid sporophytes, and the size for each of 31 chromosomes were from 0.57–2.17 μm. Besides, many DNA markers were adopted for FISH (fluorescence in situ hybridization) to S.japonica, such as sex-specific marker (Liu et al., 2012c; Gu et al., 2014), nuclear ribosomal RNA genes (Liu et al., 2017b), Arabidopsis-type telomere sequence (TTTAGGG)n (Yang et al., 2017). Recently, a total of 1 576 scaffolds, accounting for about 65% of the assembled S. japonica genome sequences, was roughly mapped to the 31 genetic linkage groups (Fan et al., 2020), and the genome of E. siliculosus was assembled into 28 linkage groups, and it was near the chromosome level, referred to the 25-chromosome number (Cormier et al., 2017). So far, using single-color FISH, only several DNA markers have been successfully anchored to the chromosomes of S. japonica, respectively. It is expected that, in the near future, Multi-color FISH, which was firstly invented to study human chromosomes (Speicher et al., 1996), could be modified and used to distinguish each of 31 tiny chromosomes with similar size.
6 MOLECULAR GENETICS FOR S. JAPONICA 6.1 Nuclear genomic studiesWith the fast development of sequencing technologies, many seaweed nuclear genomes were sequenced (Table 2). These provide the data for illustrating the structure and function of seaweed genomes and functional genes, and understanding the evolutions of multicellular algae, and unique genes related to phycocolloids and abiotic stress (Cock et al., 2010; Prochnik et al., 2010; Collén et al., 2013; Nakamura et al., 2013; Zhou et al., 2013; Ye et al., 2015; Nishitsuji et al., 2016, 2019; Brawley et al., 2017; De Clerk et al., 2018; Lee et al., 2018; Sun et al., 2018; Nishiyama et al., 2018; Arimoto et al., 2019; Liu et al., 2019). Ye et al. (2015) first reported one draft genome sequence to S. japonica, it characterized with 537 Mb from the female gametophytes of one S. japonica strain Ja, and contained 18 733 protein-coding genes annotated, it was larger than that of E. siliculosus with 16 256 genes. Compared to E.siliculosus, S. japonica process more significant gene expansion in the 58 families, which contained mainly for iodine concentration, vanadium-dependent haloperoxidases, cellulose synthase, mannuronan C-5-epimerases and alpha-(1, 6)-fructosyltransferase. Although S. japonica genome has been sequenced, assembled only at the contig and scaffold level, the gold standard reference genome without gaps still should be developed with the new generation sequencing technology at the next step. At present, many protein-coding genes have been annotated, but few were studied experimentally at the biochemistrical level (see more in part 8). Only recently small RNAs (microRNA, lncRNA) and DNA methylation were analyzed to S. japonica genome (see more in part 6.3). Noncoding functional elements such as gene-specific enhancer in the genome of S. japonica have not been explored yet.
Crépineau et al. (2000) firstly reported transcriptional analysis to L. digitata with carbon-concentrating, cell wall biosynthesis and halogen metabolism related genes. To S. japonica, it was first analyzed using EST sequencing data (Xuan et al., 2012), and more recently, RNA-seq techniques was adopted for analysis the gene transcriptional patterns related with light, temperature and growth stages (Deng et al., 2009; Wang et al., 2013a; Liu et al., 2014b; Ding et al., 2019; Shao et al., 2019a, 2019b). Under blue light conditions, 11 660 (16.5%) differentially expressed unigenes were detected compared with that in the dark treatments (Deng et al., 2009), while under 20℃, there are total of 947 up- or down-regulated genes (Liu et al., 2014b). Tanscriptomic studies for S. japonica have facilitated its genome annotation, and produced many differentially expressed genes under different conditions; however, it is still in its infancy. Integrated with the other data, such as small RNAs, proteome, to decipher the regulatory networks of S.japonica developmental biology, transcriptomic studies will be meaningful and helpful for its cultivation and breeding.
6.3 MicroRNA and DNA methylationRecently, microRNA and DNA methylation were reported in S. japonica (Liu et al., 2015; Fan et al., 2020), Cock et al. (2017) indicated that none of S. japonica miRNAs sequences are similar with that of Ectocarpus sp., which implied that miRNAs evolve rapidly in the brown algae. Fan et al. (2020) reported that the cytosine methylations are important to the formation of diploid sporophyte and haploid gametophyte life-cycle stages, tissue differentiation and metabolism. Yang et al. (2020) proved that the non-coding RNAs participate in the regulation of CRY-DASH in the growth and early development of S. japonica. These primary data showed that, compared with the land plants and animals, S. japonica has lower levels of DNA methylation, and its microRNAs play a less central role in gene regulation, which is consistent with its simple morphology and far phylogenetic distance from higher plants and animals.
6.4 Chloroplast and mitochondria genomesAt present, over 10 chloroplast genome sequences from the brown seaweeds were reported (NCBI, https://www.ncbi.nlm.nih.gov/genome/organelle/), such as E. siliculosus and Fucus vesiculosus (Le Corguillé et al., 2009), Sargassum horneri (Liu and Pang, 2016), U. pinnatifida and Costaria costata (Zhang et al., 2015d, 2015e), Dictyopteris divaricata (Liu et al., 2017a), S.confusum (Liu et al., 2018). Overall, the sizes of sequenced brown seaweed chloroplast genomes ranged from 124 kb to 140 kb, with the largest 139 954 bp for E. siliculosus (Le Corguillé et al., 2009). The G+C contents were from 28.94% to 31.19%.
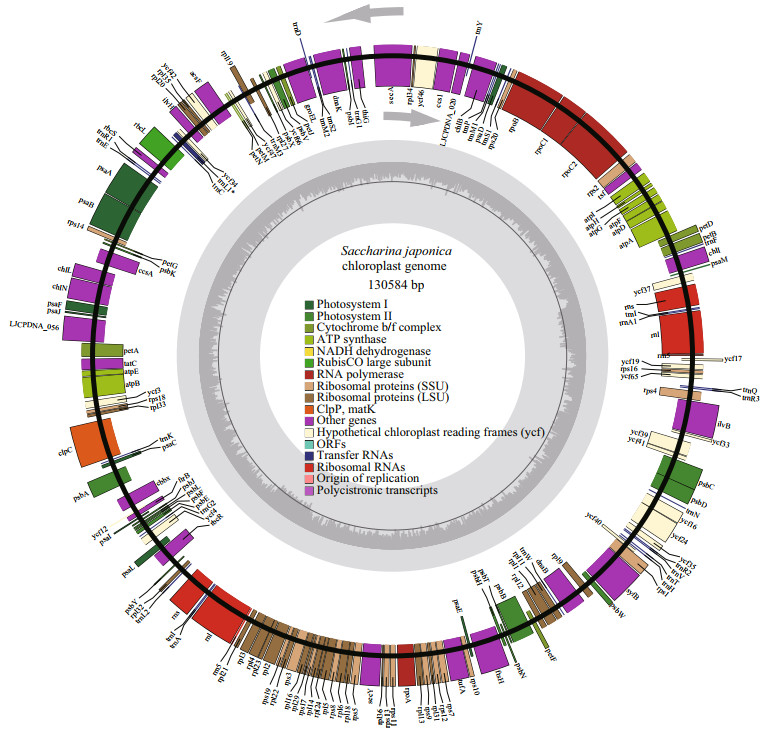
Wang et al. (2013b) firstly reported the chloroplast genome of S. japonica with 130 584 bp in size, it consisted of 139 protein-coding genes, 29 tRNA genes, and 3 ribosomal RNA genes (Fig. 2). The structure analysis showed that the S. japonica chloroplast genome has a large and a small single-copy region (LSC and SSC), separated by two copies of inverted repeats (IR1 and IR2), which is the typical structure character for the known brown seaweed chloroplast genome.
|
| Fig.2 Chloroplast genome of S.japonica (rewritten from Wang et al., 2013b by OGDRAW version 1.3.1) |
The organelles' Rubisco spacer sequences and others were applied for maternal inheritance pattern analysis in Fucus, Alaria and S. japonica (Kraan and Guiry, 2000; Coyer et al., 2002; Li et al., 2016b).
Since the report of mitochondrial genome of Pylaiella littoralis (Oudot-Le Secq et al., 2001), many other brown algal mitochondrial genomes were registered in NCBI. Generally, the size of brown algal mitochondrial genomes were from 31.6–58.5 kb, and it consisted of 3 ribosomal subunit genes (rRNAs), 24–26 transfer RNA genes (tRNAs), 35–36 protein-coding genes, and 2–16 uncharacterized open reading frames (ORFs), and some noncoding intergenic spacer regions. Yotsukura et al. (2010) analyzed the mitochondrial genomes of S. japonica, S. longipedalis, S. angustata and S. coriacea. The total size of S. japonica mitochondrial genome were 37 657 bp, which included 3 ribosomal RNA genes, 25 transfer RNA genes, 11 ribosomal small subunit protein genes and 6 large subunit genes, 10 NADH dehydrogenase genes, 3 cytochrome oxidase genes, 1 apocytochrome b gene, 3 f0-ATPase genes, 1 tatC gene and 3 ORFs.
The mitochondrial genome data were adopted for evolutional and phylogenetic studies. Liu and Pang (2015) revealed that P.fascia has a close evolutionary relationship with E. siliculosus. Balakirev et al. (2012) applied COI sequence, nuclear (ITS) and plastid (rbcLS) sequences for the morphological "longipes" (LON) and "shallow-water" (SHA) forms analysis, and indicated that the "typical" (TYP) and LON forms are genetically similar even with different phenotype and ecological variations. Moreover, the COI and trnW-trnL sequences in mitochondrial genome were either applied for phylogeographic patterns analysis in S.japonica populations (Zhang et al., 2015c), and rpl6-rps2 and trnW-trnL of mitochondrial DNA markers were adopted for maternal inheritance analysis to S. japonica (Li et al., 2016b).
7 POPULATION GENETICS AND QUANTITATIVE GENETICNeefus et al. (1993) used isozyme techniques to study Laminaria populations, and detected the lower genetic diversity within and between their populations, since then many seaweeds population genetic studies were conducted in algal biogeographical and phylogeographical analysis (Wattier and Maggs, 2001; Hu and Fraser, 2015; Zhang et al., 2015c, 2019b; Luttikhuizen et al., 2018). To the S. japonica, its cultivated population genetic diversities were lower than those in Russian and Japanese natural populations (Bi et al., 2011; Shan et al., 2011, 2017; Liu et al., 2012a; Yotsukura et al., 2016; Li et al., 2017; Zhang et al., 2017). Recently, Shan et al. (2019) proved that there are large genetic divergence in the farmed populations and the subtidal "wild" populations, and suggested that there is a need to introduce the wild S.japonica germplasm for enhancing the cultivars genetic diversity in China.
There were many traditional quantitative studies on the blade length, width and thickness, the stipe length, and the iodine content (Fang, 1983; Wu and Lin, 1987). With the integrations of feasible marker systems, QTL analysis to blade length and width were available, and usually F2 populations were used for the QTL mapping (Li et al., 2007b; Yang et al., 2009; Liu et al., 2009a, 2010a; Zhang et al., 2015b, 2015f). Wang et al. (2018) used SLAF-seq (selective length amplified fragment) techniques to generate 31 linkage groups, and localizing 12 QTLs to its blade length and 10 to width, either verified that 14 Tic20 (translocon at the inner envelope membrane of chloroplasts) and three peptidase genes are candidates related with the blade length and width. One Tic20 gene from S. japonica was verified in the diatom only (Chen et al., 2019). Compared with that in rice, where about 2 000 genes were reported to be cloned and partially studied (Li et al., 2018), although dozens of QTLs were successfully mapped especially to the blade length and width of S. japonica, few candidate genes were found to date.
8 FUNCTIONALGENEIDENTIFICATIONBefore the 21st century, for brown seaweeds, only chlorophyll a/c-binding protein fucoxanthin gene in L. saccharina (Caron et al., 1996) and one metallothionein gene in F. vesiculosus (Morris et al., 1999; Lee and Lee, 2001) were analyzed. While in S. japonica, Fu et al. (2009) firstly cloned one cDNA hsp70 full sequences and quantitatively detected its expression under the different temperature, other genes such as Rubisco gene, trehalose-6-phosphate synthase gene, AUREOCHROME gene and candidate gametophyte sex determination gene (SJHMG) were reported afterwards (Deng et al., 2014a, b; Shao et al., 2014b; Zhang et al., 2019c). Moreover, carbonic anhydrase gene, cbbX gene, mannuronan C5-epimerase gene and alginate lyase gene were either reported (Shi et al., 2010; Ye et al., 2014; Inoue et al., 2016; Inoue and Ojima, 2019). Besides, many genes were annotated after transcriptomic analysis, which related to blue and red light respectively (Deng et al., 2009; Wang et al., 2013a), under the temperature stress (Liu et al., 2014), and different developmental stages (Ding et al., 2019; Shao et al., 2019b). Biochemical characterization of mannitol-2-dehydrogenase (Shao et al., 2014a), GDP-mannose dehydrogenase genes (Zhang et al., 2016c), PMM/ PGM (phosphomannomutase/phosphoglucomutase) (Zhang et al., 2018d), CRY-DASH gene (Yang et al., 2020) were either documented. Recently, Luan et al. (2019) verified 40S ribosomal protein S6 was closely related to SjAUREO protein when detected with yeast two-hybrid library and interaction systems.
9 GENE MODIFIED TECHNIQUEThe technological platforms for gene transformation were established to crops including rice etc., especially with the Agrobacterium-mediated technology (Dunwell and Wetten, 2012). However, to develop feasible platforms for gene transformation in seaweeds is still progressing. Recently, one stable transformation was reported in U.mutabilis (Oertel et al., 2015). Mikami (2014) testified the transformation methods in seaweed. The bombardment techniques were proved to be successful in many seaweeds, such as P. yezoensis (Hirata et al., 2011, 2014; Shin et al., 2016; Kong et al., 2017), K. albariza (Wang et al., 2010), U. prolifera (Wu et al., 2018), S. horneri (Pang et al., 2019) and S. japonica (Jiang et al., 2002; Zhang et al., 2008b; Li et al., 2009), even with the disadvantages of transient expressions in the seaweeds. In general, we believed that transgenetic system could be modified with other techniques, such as RNAi, CRISPR-Cas9 manipulations, to generate one stable platform for gene transformation in S. japonica.
10 CONCLUSION AND PERSPECTIVEWith the new S. japonica genome information, we can annotate more genes with the actual functions. Nevertheless, there are many problems existed and need to be solved. Firstly, it is required to explore the function of those genes involved with alginate production and the mannitol pathway in vitro. Secondly, no genotype has been yet mapped to the phenotype of S. japonica, and it required verification of those loci related to blade length and width. Thirdly, there is an urgent need to establish one stable platform for the functional gene verification in the future. Lastly, the selection and breeding of S. japonica should be strengthened for sustainable aquaculture of commercial seaweeds in China, and the genetic and breeding strategies will be shifted from roughly to precisely in the future.
11 DATA AVAILABILITY STATEMENTThe chloroplast genome analyzed in this study has been submitted to the NCBI database with the accession number NC_018523.1. All seaweed genomic sequences were retrieved from NCBI genome database.
12 ACKNOWLEDGMENTWe thank anonymous reviewers for their critical comments and suggestions for this paper, and Dr. Alan T Critchley for the help in English revision.
Arimoto A, Nishitsuji K, Higa Y, Arakaki N, Hisata K, Shinzato C, Satoh N, Shoguchi E. 2019. A siphonous macroalgal genome suggests convergent functions of homeobox genes in algae and land plants. DNA Research, 26(2): 183-192.
DOI:10.1093/dnares/dsz002 |
Balakirev E S, Krupnova T N, Ayala F J. 2012. DNA variation in the phenotypically-diverse brown alga Saccharina japonica. BMC Plant Biology, 12(1): 108.
DOI:10.1186/1471-2229-12-108 |
Bartsch I, Wiencke C, Bischof K, Buchholz C M, Buck B H, Eggert A, Feuerpfeil P, Hanelt D, Jacobsen S, Karez R, Karsten U, Molis M, Roleda M Y, Schubert H, Schumann R, Valentin K, Weinberger F, Wiese J. 2008. The genus Laminaria sensu lato: recent insights and developments. European Journal of Phycology, 43(1): 1-86.
DOI:10.1080/09670260701711376 |
Bi Y H, Hu Y J, Zhou Z G. 2011. Genetic variation of Laminaria japonica (Phaeophyta) populations in China as revealed by RAPD markers. Acta Oceanologica Sinica, 30(2): 103-112.
DOI:10.1007/s13131-011-0110-y |
Billot C, Rousvoal S, Estoup A, Epplen J T, Saumitou-Laprade P, Valero M, Kloareg B. 1998. Isolation and characterization of microsatellite markers in the nuclear genome of the brown alga Laminaria digitata(Phaeophyceae). Molecular Ecology, 7(12): 1 778-1 780.
DOI:10.1046/j.1365-294x.1998.00516.x |
Brawley S H, Blouin N A, Ficko-Blean E, Wheeler G L, Lohr M, Goodson H V, Jenkins J W, Blaby-Haas C E, Helliwell K E, Chan C X, Marriage T N, Bhattachaya D, Klein A S, Badis Y, Brodie J, Cao Y Y, Collén J, Dittami S M, Gachon C M M, Green B R, Karpowicz S J, Kim J W, Kudahl U J, Lin S J, Michel G, Mittag M, Olsen B J S C, Pangilinan J L, Peng Y, Qiu H, Shu S Q, Singer J T, Smith A G, Sprecher B N, Wagner V, Wang W F, Wang Z Y, Yan J Y, Yarish C, Zäuner-Riek S, Zhuang Y Y, Zou Y, Lindquist E A, Grimwood J, Barry K W, Rokhsar D S, Schmutz J, Stiler J W, Grossman A R, Prochnik S E. 2017. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proceedings of the National Academy of Sciences of the United States of America, 114(31): E6361-E6370.
DOI:10.1073/pnas.1703088114 |
Caron L, Douady D, Quinet-Szely M, de Goër S, Berkaloff C. 1996. Gene structure of a chlorophyll a/c-binding protein from a brown alga: presence of an intron and phylogenetic implications. Journal of Molecular Evolution, 43(3): 270-280.
DOI:10.1007/BF02338835 |
Chen Z H, Wang X L, Li S, Yao J T, Shao Z R, Duan D L. 2019. Verification of the Saccharina japonica translocon Tic20 and its localization in the chloroplast membrane in diatoms. International Journal of Molecular Sciences, 20: 4 000.
DOI:10.3390/ijms20164000 |
Cock J M, Liu F L, Duan D L, Bourdareau S, Lipinska A P, Coelho S M, Traver J E. 2017. Rapid evolution of microRNA loci in the brown algae. Genome Biology and Evolution, 9(3): 740-749.
DOI:10.1093/gbe/evx038 |
Cock J M, Sterck L, Rouzé P, Scornet D, Allen A E, Amoutzias G, Anthouard V, Artiguenave F, Aury J M, Badger J H, Beszter B, Billiau K, Bonnet E, Bothwell J H, Bowler C, Boyen C, Brownlee C, Carrano C J, Charrier B, Cho G Y, Coelho S M, Collén J, Corre E, Da Silva C, Delage L, Delaroque N, Dittami S M, Doulbeau S, Elias M, Farnham G, Gachon C M M, Gschloessl B, Heesch S, Jabbar K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper F C, Lang D, Le Bail A, Leblanc C, Lerouge P, Lohr M, Lopez P J, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli C A, Nelson D R, Nyvall-Collén P, Peters A F, Pommier C, Potin P, Poulain J, Quesneville H, Read B, Rensing S A, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder D C, Ségurens B, Strittmatter M, Tonon T, Tregear J W, Valentin K, von Dassow P, Yamagishi T, van der Peer Y, Wincker P. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature, 465(7298): 617-621.
DOI:10.1038/nature09016 |
Collén J, Porcel B, Carré W, Ball S G, Chaparo C, Tonon T, Barbeyron T, Michel G, Noel B, Valentin K, Elias M, Artiguenave F, Arun A, Aury J M, Barbosa-Neto J F, Bothwell J H, Bouget F Y, Brillet L, Cabello-Hurtado F, Capella-Gutiérrez S, Charrier B, Cladière L, Cock J M, Coelho S M, Colleoni C, Czjzek M, Da Silva C, Delage L, Denoeud F, Deschamps P, Dittami S M, Gabaldón T, Gachon C M M, Groisillier A, Hervé C, Jabbari K, Katinka M, Kloareg B, Kowalczyk N, Labadie K, Leblanc C, Lopez P J, McLachlan D H, Meslet-Cladiere L, Moustafa A, Nehr Z, Collén P N, Panaud O, Partensky F, Poulain J, Rensing S A, Rousvoal S, Samson G, Symeonidi A, Weissenbach J, Zambounis A, Wincker P, Boyen C. 2013. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proceedings of the National Academy of Sciences of the United States of America, 110(13): 5 247-5 252.
DOI:10.1073/pnas.1221259110 |
Cormier A, Avia K, Sterck L, Derrien T, Wucher V, Andres G, Monsoor M, Godfroy O, Lipinska A, Perrineau M M, van De Peer Y, Hitte C, Corre E, Coelho S M, Cock J M. 2017. Re-annotation, improved large-scale assembly and establishment of a catalogue of noncoding loci for the genome of the model brown alga Ectocarpus. New Phytologist, 214(1): 219-232.
DOI:10.1111/nph.14321 |
Coyer J A, Peters A F, Hoarau G, Staw W T, Olsen J L. 2002. Inheritance paterns of ITS1, chloroplasts and mitochondria in artificial hybrids of the seaweeds Fucus serratus and F.evanescens (Phaeophyceae). European Journal of Phycology, 37(2): 173-178.
DOI:10.1017/S0967026202003682 |
Crépineau F, Roscoe T, Kaas R, Kloareg B, Boyen C. 2000. Characterisation of complementary DNAs from the expressed sequence tag analysis of life cycle stages of Laminaria digitata (Phaeophyceae). Plant Molecular Biology, 43(4): 503-513.
DOI:10.1023/A:1006489920808 |
De Clerck O, Kao S M, Bogaert K A, Blomme J, Foflonker F, Kwantes M, Vancaester E, Vanderstraeten L, Aydogdu E, Boesger J, Califano G, Charrier B, Clewes R, Del Cortona A, D'Hondt S, Fermandez-Pozo N, Gachon C M, Hanikenne M, Lattermann L, Leliaert F, Liu X J, Maggs C A, Popper Z A, Raven J A, van Bel M, Wilhelmsson P K I, Bhattacharya D, Coates J C, Rensing S A, van der Straeten D, Vardi A, Sterck L, Vandepoele K, van de Peer Y, Wichard T, Bothwell J H. 2018. Insights into the evolution of multicellularity from the sea lettuce genome. Current Biology, 28(18): 2 921-2 933.e5.
DOI:10.1016/j.cub.2018.08.015 |
Deng Y Y, Wang X L, Guo H, Duan D L. 2014b. A trehalose-6-phosphate synthase gene from Saccharina japonica(Laminariales, Phaeophyta). Molecular Biology Reports, 41(1): 529-536.
DOI:10.1007/s11033-013-2888-5 |
Deng Y Y, Yao J T, Fu G, Guo H, Duan D L. 2014a. Isolation, expression, and characterization of blue light receptor AUREOCHROME gene from Saccharina japonica(Laminariales, Phaeophyta). Marine Biotechnology, 16(2): 135-143.
DOI:10.1007/s10126-013-9539-7 |
Deng Y Y, Yao J T, Wang X L, Guo H, Duan D L. 2009. Transcriptome sequencing and comparative analysis of Saccharina japonica (Laminariales, Phaeophyceae) under blue light induction. PLoS One, 7(6): e39704.
DOI:10.1371/journal.pone.0039704 |
Ding H Y, Guo L, Li X J, Yang G P. 2019. Transcriptome analysis of kelp Saccharina japonica unveils its weird transcripts and metabolite shift of main components at different sporophyte developmental stages. Journal of Oceanology and Limnology, 37(2): 640-650.
DOI:10.1007/s00343-019-8019-y |
Duan D L, Miao G R, Wang X L. 2015. Aquacultural Biology of Saccharina japonica. Scientific press, Beijing.
(in Chinese)
|
Dunwell J M, Wetten A C. 2012. Transgenic Plants: Methods and Protocols. 2nd edn. Human Press, Totowa, NJ.
|
Fan X, Han W T, Teng L H, Jiang P, Zhang X W, Xu D, Li C, Pellegrini M, Wu C H, Wang Y T, Kaczurowski M J S, Lin X, Tirichine L, Mock T, Ye N H. 2020. Single-base methylome profiling of the giant kelp Saccharina japonica reveals significant differences in DNA methylation to microalgae and plants. New Phytologist, 225(1): 234-249.
DOI:10.1111/nph.16125 |
Fang T C, Cui J J, Ou Y L, Dai J X, Wang M L, Liu Q S, Yang Q M. 1983. Breeding of the new variety "DanHai No.1" of Laminaria japonica by using a female haploid clone of the kelp. Journal of Shandong College of Oceanology, 13(4): 63-70.
(in Chinese with English abstract) |
Fang T C, Jiang B Y, Li J J. 1965. Further studies of the genetics of Laminaria frond-length. Oceanologia et Limnologia Sinica, 7(1): 59-66.
(in Chinese with English abstract) |
Fang T C, Ou Y L, Cui J J. 1985. Breeding of hybrid Laminaria "DanZa No.10"-an application of the Laminarian haploid cell clones. Journal of Shandong College of Oceanology, 15(1): 64-72.
(in Chinese with English abstract) |
Fang T C, Wu C Y, Jiang B Y, Li J J, Ren G Z. 1963. The breeding of a new variety of Haidai (Laminaria japonica Aresch.). Science in China, Series A, 12(7): 1 011-1 018.
|
Fang T C, Zhang D M. 1982. Mr. Otuki and the early history of mariculture of Laminaria japonica in China. Journal of Shandong College of Oceanology, 12(3): 97-98.
(in Chinese with English abstract) |
Fang T C. 1983. Genetic studies for Laminaria japonica in China. Acta Oceanologica Sinica, 5(4): 500-506.
(in Chinese) |
FAO. 2018. The state of world fisheries and aquaculture.Rome, Italy, http://www.fao.org/3/i9540en/I9540EN.pdf.
|
Fu W D, Yao J T, Wang X L, Liu F L, Fu G, Duan D L. 2009. Molecular cloning and expression analysis of a cytosolic Hsp70 gene from Laminaria japonica (Laminariaceae, Phaeophyta). Marine Biotechnology, 11(6): 738-747.
DOI:10.1007/s10126-009-9188-z |
Gu J G, Sun Y P, Liu Y, Bi Y H, Zhou Z G. 2014. Sex identification and genetic variation of Saccharina(Phaeophyta) gametophytes as revealed by inter-simple sequence repeat (ISSR) markers. Journal of Applied Phycology, 26(1): 635-646.
DOI:10.1007/s10811-013-0089-1 |
Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1. 3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research, 47: W59-W64.
|
Hafting J T, Craigie J S, Stengel D B, Loureiro R R, Buschmann A H, Yarish C, Edwards M D, Critchley A T. 2015. Prospects and challenges for industrial production of seaweed bioactives. Journal of Phycology, 51(5): 821-837.
DOI:10.1111/jpy.12326 |
Hirata R, Takahashi M, Saga N, Mikami K. 2011. Transient gene expression system established in Porphyra yezoensis is widely applicable in Bangiophycean algae. Marine Biotechnology, 13(5): 1 038-1 047.
DOI:10.1007/s10126-011-9367-6 |
Hirata R, Uji T, Fukuda S, Mizuta H, Fujiyama A, Tabata S, Saga N. 2014. Development of a nuclear transformation system with a codon-optimized selection marker and reporter genes in Pyropia yezoensis (Rhodophyta). Journal of Applied Phycology, 26(4): 1 863-1 868.
DOI:10.1007/s10811-013-0234-x |
Hu Z M, Fraser C. 2015. Seaweed Phylogeography: Adaptation and Evolution of Seaweeds under Environmental Change. Springer Science and Business Media, Dordrecht.
|
Hwang E K, Yotsukura N, Pang S J, Shan T F. 2019. Seaweed breeding programs and progress in eastern Asian countries. Phycologia, 58(5): 484-495.
DOI:10.1080/00318884.2019.1639436 |
Inoue A, Ojima T. 2019. Functional identification of alginate lyase from the brown alga Saccharina japonica. Scientific Reports, 9(1): 4 937.
DOI:10.1038/s41598-019-41351-6 |
Inoue A, Satoh A, Morishita M, Tokunaga Y, Miyakawa T, Tanokura M, Ojima T. 2016. Functional heterologous expression and characterization of mannuronan C5-epimerase from the brown alga Saccharina japonica. Algal Research, 16: 282-291.
DOI:10.1016/j.algal.2016.03.030 |
Jiang P, Qin S, Tseng C K. 2002. Expression of hepatitis B surface antigen gene (HBsAg) in Laminaria japonica(Laminariales, Phaeophyta). Chinese Science Bulletin, 47(17): 1 438-1 440.
DOI:10.1360/02tb9317 |
Kong F N, Zhao H L, Liu W X, Li N, Mao Y X. 2017. Construction of plastid expression vector and development of genetic transformation system for the seaweed Pyropia yezoensis. Marine Biotechnology, 19(2): 147-156.
DOI:10.1007/s10126-017-9736-x |
Kraan S, Guiry M D. 2000. Molecular and morphological character inheritance in hybrids of Alaria esculenta and A. praelonga (Alariaceae, Phaeophyceae). Phycologia, 39(6): 554-559.
DOI:10.2216/i0031-8884-39-6-554.1 |
Lane C E, Mayes C, Druehl L D, Saunders G W. 2006. A multigene molecular investigation of the kelp (Laminariales, Phaeophyceae) supports substantial taxonomic reorganization. Journal of Phycology, 42(2): 493-512.
DOI:10.1111/j.1529-8817.2006.00204.x |
Le Corguillé G, Pearson G, Valente M, Viegas C, Gschloess B, Corre E, Bailly X, Peters A F, Jubin C, Vacherie B, Cock J M, Leblanc C. 2009. Plastid genomes of two brown algae, Ectocarpus siliculosus and Fucus vesiculosus:further insights on the evolution of red-algal derived plastids. BMC Evolutionary Biology, 9(1): 253.
DOI:10.1186/1471-2148-9-253 |
Lee J M, Yang E C, Graf L, Yang J H, Qiu H, Zelzion U, Chan C X, Stephens T G, Weber A P M, Boo G H, Boo S M, Kim K M, Shin Y, Jung M, Lee S J, Yim H S, Lee J H, Bhattachara D, Yoon H S. 2018. Analysis of the draft genome of the red seaweed Gracilariopsis chorda provides insights into genome size evolution in Rhodophyta. Molecular Biology and Evolution, 35(8): 1 869-1 886.
DOI:10.1093/molbev/msy081 |
Lee Y K, Lee H K. 2001. Eukaryotic algal genes and progress in molecular biology of eukaryotic algae. Algae, 16(1): 1-19.
|
Lewis R J, Jiang B Y, Neushul M, Fei X G. 1993. Hapoid parthenogenetic sporophytes of Laminaria japonica(Phaeophyceae). Journal of Phycology, 29(3): 363-369.
DOI:10.1111/j.0022-3646.1993.00363.x |
Lewis R J. 1996. Chromosomes of the brown algae. Phycologia, 35(1): 19-40.
DOI:10.2216/i0031-8884-35-1-19.1 |
Li D P, Zhou Z G, Liu H H, Wu C Y. 1999. A new method of Laminaria japonica strain selection and sporeling raising by the use of gametophyte clones. Hydrobiologia, 398-399: 473-476.
DOI:10.1023/A:1017090130586 |
Li F C, Qin S, Jiang P, Wu Y, Zhang W. 2009. The integrative expression of GUS gene driven by FCP promoter in the seaweed Laminaria japonica (Phaeophyta). Journal of Applied Phycology, 21(3): 287-293.
DOI:10.1007/s10811-008-9366-9 |
Li Q Y, Wang X L, Zhang J, Yao J T, Duan D L. 2016b. Maternal inheritance of organellar DNA demonstrated with DNA markers in crosses of Saccharina japonica(Laminariales, Phaeophyta). Journal of Applied Phycology, 28(3): 2 019-2 026.
DOI:10.1007/s10811-015-0687-1 |
Li Q Y, Zhang J, Yao J T, Wang X L, Duan D L. 2016a. Development of Saccharina japonica genomic SSR markers using next-generation sequencing. Journal of Applied Phycology, 28(2): 1 387-1 390.
DOI:10.1007/s10811-015-0643-0 |
Li T, Liu F L, Wang F J, Sun X T, Wang W J. 2012. The genetic analysis and evaluate of Saccharina Huangguan No.1. Journal of Qingdao Agricultural University, 29(3): 212-217.
(in Chinese with English abstract) |
Li X J, Cong Y Z, Yang G P, Shi Y Y, Qu S C, Wang G W, Zhang Z Z, Luo S J, Dai H L, Xie J Z, Jiang G L, Liu J L, Wang T Y. 2007a. Trait evaluation and trial cultivation of Dongfang No. 2, the hybrid of a male gametophyte clone of Laminaria longissima (Laminariales, Phaeophyta) and a female one of L. japonica. Journal of Applied Phycology, 19(2): 139-151.
DOI:10.1007/s10811-006-9120-0 |
Li X J, Liu J L, Cong Y Z, Qu S C, Zhang Z Z, Dai H L, Luo S J, Han X B, Huang S S, Wang Q Y, Liang G J, Sun J, Jin Y, Wang D Q, Yang G P. 2008a. Breeding and trial cultivation of Dongfang No.3, a hybrid of Laminaria gametophyte clones with a more than intraspecific but less than interspecific relationship. Aquaculture, 280(1-4): 76-80.
DOI:10.1016/j.aquaculture.2008.05.005 |
Li X J, Yang G P, Shi Y Y, Cong Y Z, Che S, Qu S C, Li Z L. 2008b. Prediction of the heterosis of Laminaria hybrids with the genetic distance between their parental gametophyte clones. Journal of Applied Phycology, 20(6): 1 097-1 102.
DOI:10.1007/s10811-008-9321-9 |
Li X J, Zhang Z Z, Qu S C, Liang G J, Sun J, Zhao N, Cui C J, Cao Z M, Li Y, Pan J H, Yu S H, Wang Q Y, Li X, Luo S J, Song S F, Guo L, Yang G P. 2016c. Improving seedless kelp (Saccharina japonica) during its domestication by hybridizing gametophytes and seedling-raising from sporophytes. Scientific Reports, 6(1): 21255.
DOI:10.1038/srep21255 |
Li X J, Zhang Z Z, Qu S C, Liang G J, Zhao N, Sun J, Song S F, Cao Z M, Li X, Pan J H, Luo S J, Zhang L, Cui C J, Peng J, Li Y, Wu R N, Zhao J P, Qian R, Wang L Q, Sai S, Yang G P. 2016d. Breeding of an intraspecific kelp hybrid Dongfang No.6 (Saccharina japonica, Phaeophyceae, Laminariales) for suitable processing products and evaluation of its culture performance. Journal of Applied Phycology, 28(1): 439-447.
DOI:10.1007/s10811-015-0562-0 |
Li X, Pang S J, Shan T F. 2017. Genetic diversity and population structure among cultivars of Saccharina japonica currently farmed in northern China. Phycological Research, 65(2): 111-117.
DOI:10.1111/pre.12167 |
Li Y H, Yang Y X, Liu J D, Wang X L, Gao T X, Duan D L. 2007b. Genetic mapping of Laminaria japonica and L. longissima using amplified fragment length polymorphism markers in a "two-way pseudo-testcross" strategy. Journal of Integrative Plant Biology, 49(3): 392-400.
DOI:10.1111/j.1744-7909.2007.00397.x |
Li Y, Xiao J H, Chen L L, Huang X H, Cheng Z K, Han B, Zhang Q F, Wu C Y. 2018. Rice functional genomics research: past decade and future. Molecular Plant, 11(3): 359-380.
DOI:10.1016/j.molp.2018.01.007 |
Liu F L, Shao Z R, Zhang H N, Liu J D, Wang X L, Duan D L. 2010a. QTL mapping for frond length and width in Laminaria japonica Aresch (Laminarales, Phaeophyta)using AFLP and SSR markers. Marine Biotechnology, 12(4): 386-394.
DOI:10.1007/s10126-009-9229-7 |
Liu F L, Sun X T, Wang F J, Wang W J, Liang Z R, Lin Z L, Dong Z A. 2014a. Breeding, economic traits evaluation, and commercial cultivation of a new Saccharina variety "Huangguan No.1". Aquaculture International, 22(5): 1 665-1 675.
DOI:10.1007/s10499-014-9772-8 |
Liu F L, Wang F J, Duan D L. 2012b. EST-SSR markers derived from Laminaria digitata and its transferable application in Saccharina japonica. Journal of Applied Phycology, 24(3): 501-505.
DOI:10.1007/s10811-012-9807-3 |
Liu F L, Wang W J, Sun X T, Liang Z R, Wang F J. 2014b. RNA-seq revealed complex response to heat stress on transcriptomic level in Saccharina japonica (Laminariales, Phaeophyta). Journal of Applied Phycology, 26(3): 1 585-1 596.
DOI:10.1007/s10811-013-0188-z |
Liu F L, Wang W J, Sun X T, Liang Z R, Wang F J. 2015. Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant, Cell & Environment, 38(7): 1 357-1 367.
DOI:10.1111/pce.12484 |
Liu F L, Wang X L, Liu J D, Fu W D, Duan D L, Yang Y X. 2009a. Genetic mapping of the Laminaria japonica(Laminarales, Phaeophyta) using amplified fragment length polymorphism markers. Journal of Phycology, 45(5): 1 228-1 233.
DOI:10.1111/j.1529-8817.2009.00729.x |
Liu F L, Wang X L, Yao J T, Fu W D, Duan D L. 2010b. Development of expressed sequence tag-derived microsatellite markers for Saccharina (Laminaria)japonica. Journal of Applied Phycology, 22(1): 109-111.
DOI:10.1007/s10811-009-9426-9 |
Liu F L, Yao J T, Wang X L, Hu Z M, Duan D L. 2011. Identification of SCAR marker linking to longer frond length of Saccharina japonica (Laminariales, Phaeophyta)using bulked-segregant analysis. Journal of Applied Phycology, 23(4): 709-713.
DOI:10.1007/s10811-010-9567-x |
Liu F L, Yao J T, Wang X L, Repnikova A, Galanin D A, Duan D L. 2012a. Genetic diversity and structure within and between wild and cultivated Saccharina japonica(Laminariales, Phaeophyta) revealed by SSR markers. Aquaculture, 358-359: 139-145.
DOI:10.1016/j.aquaculture.2012.06.022 |
Liu F, Jin Z, Wang Y, Bi Y P, Melton J T. 2017a. Plastid genome of Dictyopteris divaricata (Dictyotales, Phaeophyceae):understanding the evolution of plastid genomes in brown algae. Marine Biotechnology, 19(6): 627-637.
DOI:10.1007/s10126-017-9781-5 |
Liu F, Pan J, Zhang Z S, Moejes F W. 2018. Organelle genomes of Sargassum confusum (Fucales, Phaeophyceae):mtDNA vs cpDNA. Journal of Applied Phycology, 30(4): 2 715-2 722.
DOI:10.1007/s10811-018-1461-y |
Liu F, Pang S J. 2015. Mitochondrial phylogenomics reveals a close relationship between Petalonia fascia(Scytosiphonaceae, Phaeophyceae) and Ectocarpus siliculosus. Journal of Applied Phycology, 27(2): 1 021-1 028.
DOI:10.1007/s10811-014-0386-3 |
Liu F, Pang S J. 2016. Chloroplast genome of Sargassum horneri (Sargassaceae, Phaeophyceae): comparative chloroplast genomics of brown algae. Journal of Applied Phycology, 28(2): 1 419-1 426.
DOI:10.1007/s10811-015-0609-2 |
Liu T, Wang X M, Wang G L, Jia S G, Liu G M, Shan G L, Chi S, Zhang J, Yu Y H, Xue T, Yu J. 2019. Evolution of complex thallus alga: genome sequencing of Saccharina japonica. Frontiers in Genetics, 10: 378.
DOI:10.3389/fgene.2019.00378 |
Liu Y S, Li L H, Wu W K, Zhou Z G. 2009b. A SCAR molecular marker specifically related to the female gametophytes of Saccharina (Laminaria) japonica (Phaeophyta). Journal of Phycology, 45(4): 894-897.
DOI:10.1111/j.1529-8817.2009.00719.x |
Liu Y, Bi Y H, Gu J G, Li L H, Zhou Z G. 2012c. Localization of a female-specific marker on the chromosomes of the brown seaweed Saccharina japonica using fluorescence in situ hybridization. PLoS One, 7(11): e48784.
DOI:10.1371/journal.pone.0048784 |
Liu Y, Bi Y H, Zhou Z G. 2012d. Karyological observation on Saccharina japonica chromosomes stained with DAPI. Journal of Fisheries of China, 36(1): 50-54.
(in Chinese with English abstract) DOI:10.3724/SP.J.1231.2012.27590 |
Liu Y, Yang Q F, Dong W S, Bi Y H, Zhou Z G. 2017b. Characterization and physical mapping of nuclear ribosomal RNA (rRNA) genes in the haploid gametophytes of Saccharina japonica (Phaeophyta). Journal of Applied Phycology, 29(5): 2 695-2 706.
DOI:10.1007/s10811-017-1206-3 |
Luan H X, Yao J T, Chen Z H, Duan D L. 2019. The 40s ribosomal protein s6 response to blue light by interaction with SjAUREO in Saccharina japonica. International Journal of Molecular Sciences, 20(10): 2 414.
DOI:10.3390/ijms20102414 |
Luttikhuizen P C, van den Heuvel F H M, Rebours C, Witte H J, van Bleijswijk J D L, Timmermans K. 2018. Strong population structure but no equilibrium yet: Genetic connectivity and phylogeography in the kelp Saccharina latissima (Laminariales, Phaeophyta). Ecology and Evolution, 8(8): 4 265-4 277.
DOI:10.1002/ece3.3968 |
Mikami K. 2014. A technical breakthrough close at hand:feasible approaches toward establishing a gene-targeting genetic transformation system in seaweeds. Frontier in Plant Science, 5: 498.
DOI:10.3389/fpls.2014.00498 |
Morris C A, Nicolaus B, Sampson V, Harwood J L, Kille P. 1999. Identification and characterization of a recombinant metallothionein protein from a marine alga, Fucus vesiculosus. Biochemical Journal, 338(2): 553-560.
DOI:10.1042/bj3380553 |
Nakahara H. 1984. Alternation of generations of some brown algae in unialgal and axenic cultures. Scientific Papers of the Institute of Algological Research, Hokkaido University, 7(2): 77-194.
|
Nakamura Y, Sasaki N, Kobayashi M, Ojima N, Yasuike M, Shigenobu Y, Satomi M, Fukuma Y, Shiwaku K, Tsujimoto A, Kobayashi T, Nakayama I, Ito F, Nakajima K, Sano M, Wada T, Kuhara S, Inouye K, Gojobori T, Ikeo K. 2013. The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS One, 8(3): e57122.
DOI:10.1371/journal.pone.0057122 |
Neefus C D, Allen B P, Baldwin H P, Mathieson A C, Eckert R T, Yarish C, Miller M A. 1993. An examination of the population genetics of Laminaria and other brown algae in the Laminariales using starch gel electrophoresis. Hydrobiologia, 260-261: 67-79.
DOI:10.1007/BF00049005 |
Nishitsuji K, Arimoto A, Higa Y, Mekaru M, Kawamitus M, Satoh N, Shoguchi E. 2019. Draft genome of the brown alga, Nemacystus decipiens, Onna-1 strain: fusion of genes involved in the sulfated fucan biosynthesis pathway. Scientific Reports, 9: 4 607.
DOI:10.1038/s41598-019-40955-2 |
Nishitsuji K, Arimoto A, Iwai K, Sudo Y, Hisata K, Fujie M, Arakaki N, Kushiro T, Konish T, Shinzato C, Satoh N, Shoguchi E. 2016. A draft genome of the brown alga, Cladosiphon okamuranus, S-strain: a platform for future studies of 'mozuku' biology. DNA Research, 23(6): 561-570.
DOI:10.1093/dnares/dsw039 |
Nishiyama T, Sakayama H, de Vries J, Bushchmann H, SaintMarcoux D, Ullrich K K, Haas F B, Vanderstraeten L, Becker D, Lang D, Vosolsobë, Rombauts S, Wihelmsson P K I, Janitza P, Kem R, Heyl A, Rümpler F, Villalobos L A C, Clay J M, Skokan R, Toyoda A, Suzuki Y, Kagoshima H, Schijlen E, Tajeshwar N, Catarino B, Hetherington A J, Saltykova A, Bonnot C, Breuninger H, Symeonidi A, Radhakrishnan G V, van Nieuwerburgh F, Deforce D, Chang C, Karol K G, Hedrich R, Ulvskov P, Glöckner G, Delwiche C F, Petrášek J, van de Peer Y, Friml J, Beilby M, Dolan L, Kohara Y, Sugano S, Fujiyama A, Delaux P M, Quint M, Theißen G, Hagemann M, Harholt J, Dunand C, Zachgo S, Langdale J, Maumus F, Van Der Straeten D, Gould S B, Rensing S A. 2018. The Chara genome:secondary complexity and implications for plant terretrialization. Cell, 174(2): 448-464.e24.
DOI:10.1016/j.cell.2018.06.033 |
Oertel W, Wichard T, Weissgerber A. 2015. Transformation of Ulva mutabilis (Chlorophyta) by vector plasmids integrating into the genome. Journal of Phycology, 51(5): 963-979.
DOI:10.1111/jpy.12336 |
Oudot-Le Secq M P, Fontaine J M, Rousvoal S, Kloareg B, Loiseaux-de Goër S. 2001. The complete sequence of a brown algal mitochondrial genome, the Ectocarpale Pylaiella littoralis (L. ) Kjellm. Journal of Molecular Evolution, 53(2): 80-88.
DOI:10.1007/s002390010196 |
Pang S J, Liu F, Liu Q S, Wang J Q, Sun C B. 2015. Breeding and genetic stability evaluation of the new Saccharina variety "205". China Fisheries, (10): 59-60.
(in Chinese) |
Pang S J, Wu C Y. 1996. Study on gametophyte vegetative growth of Undaria pinnatifida and its applications. Chinese Journal of Oceanology and Limnology, 14(3): 205-210.
DOI:10.1007/BF02850381 |
Pang Y L, Li Y, Liu Z Y, Cui Y L, Qin S. 2019. Transient expression of the enhanced green fluorescent protein(egfp) gene in Sargassum horneri. Journal of Oceanology and Limnology, 37(2): 651-656.
DOI:10.1007/s00343-019-8014-3 |
Patwary M, van der Meer J P. 1992. Genetics and breeding of cultivated seaweeds. The Korea Journal of Phycology, 7(2): 281-318.
|
Peng J, Zhang L, Li X J, Cui C J, Wu R N, Tian P P, Li Y, Liu Y L. 2016. Development of genic SSR markers from an assembled Saccharina japonica genome. Journal of Applied Phycology, 28(4): 2 479-2 484.
DOI:10.1007/s10811-015-0747-6 |
Prochnik S E, Umen J, Nedelcu A M, Hallman A, Miller S M, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin L K, Hellsten U, Chapman J, Simakov O, Rensing S A, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev I V, Schmitt R, Kirk D, Rokhsar D S. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science, 329(5988): 223-226.
DOI:10.1126/science.1188800 |
Selivanova O N, Levenetz I R, Ogorodnikov V S. 2006.Seaweed resources of the far east of Russia. In: Seaweed resources of the world, Critchley A T, Ohno M, Largo D, et al. eds.
|
Shan T F, Li Q X, Wang X M, Su L, Pang S J. 2019. Assessment of the genetic connectivity between farmed populations on a typical kelp farm and adjacent spontaneous populations of Saccharina japonica (Phaeophyceae, Laminariales) in China. Frontiers in Marine Science, 6: 494.
DOI:10.3389/fmars.2019.00494 |
Shan T F, Pang S J, Zhang Y R, Yakovleva I M, Skriptsova A V. 2011. An AFLP-based survey of genetic diversity and relationships of major farmed cultivars and geographically isolated wild populations of Saccharina japonica(Phaeophyta) along the northwest coasts of the pacific. Journal of Applied Phycology, 23(1): 35-45.
DOI:10.1007/s10811-010-9530-x |
Shan T F, Yotsukura N, Pang S J. 2017. Novel implications on the genetic structure of representative populations of Saccharina japonica (Phaeophyceae) in the Northwest Pacific as revealed by highly polymorphic microsatellite markers. Journal of Applied Phycology, 29(1): 631-638.
DOI:10.1007/s10811-016-0888-2 |
Shao Z R, Liu F L, Li Q Y, Yao J T, Duan D L. 2014b. Characterization of ribulose-1, 5-bisphosphate carboxylase/oxygenase and transcriptional analysis of its related genes in Saccharina japonica (Laminariales, Phaeophyta). Chinese Journal of Oceanology and Limnology, 32(2): 377-389.
DOI:10.1007/s00343-014-3130-6 |
Shao Z R, Wang W L, Zhang P Y, Yao J T, Wang F H, Duan D L. 2019a. Genome-wide identification of genes involved in carbon fixation in Saccharina japonica and responses of putative C4-related genes to bicarbonate concentration and light intensity. Plant Physiology & Biochemistry, 137: 75-83.
DOI:10.1016/j.plaphy.2019.01.032 |
Shao Z R, Zhang P Y, Li Q Y, Wang X L, Duan D L. 2014a. Characterization of mannitol-2-dehydrogenase in Saccharina japonica: evidence for a new polyol-specific long-chain dehydrogenases/reductase. PLoS One, 9(5): e97935.
DOI:10.1371/journal.pone.0097935 |
Shao Z R, Zhang P Y, Lu C, Li S X, Chen Z H, Wang X L, Duan D L. 2019b. Transcriptome sequencing of Saccharina japonica sporophytes during whole developmental periods reveals regulatory networks underlying alginate and mannitol biosynthesis. BMC Genomics, 20(1): 975.
DOI:10.1186/s12864-019-6366-x |
Shendure J, Ji H. 2008. Next-generation DNA sequencing. Nature Biotechnology, 26(10): 1 135-1 145.
DOI:10.1038/nbt1486 |
Shi W W, Wang L L, Chen J, Ouyang L L, Zhou Z G. 2010. Characterization and differential expression of cbbX gene between female and male gametophytes of Laminaria japonica. Journal of Fisheries of China, 34(1): 80-88.
(in Chinese with English abstract) DOI:10.3724/SP.J.1231.2010.06456 |
Shi Y Y, Yang G P, Liao M J, Li X J, Cong Y Z, Qu S C, Wang T Y. 2008. Parentage analysis of Dongfang No. 2, a hybrid of a female gametophyte clone of Laminaria japonica(Laminariales, Phaeophyta) and a male clone of L.longissima. Journal of Ocean University of China, 7(2): 193-198.
DOI:10.1007/s11802-008-0193-z |
Shi Y Y, Yang G P, Liu Y J, Liao M J, Cong Y Z. 2007. Development of 18 polymorphic microsatellite DNA markers of Laminaria japonica (Phaeophyceae). Molecular Ecology Notes, 7(4): 620-622.
DOI:10.1111/j.1471-8286.2006.01652.x |
Shin Y J, Lim J M, Park J H, Choi D W, Hwang M S, Park E J, Min S R, Kim S W, Jeong W J. 2016. Characterization of PyGUS gene silencing in the red macroalga, Pyropia yezoensis. Plant Biotechnology Reports, 10(6): 359-367.
DOI:10.1007/s11816-016-0408-5 |
Speicher M R, Ballard S G, Ward D C. 1996. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nature Genetics, 12(4): 368-375.
DOI:10.1038/ng0496-368 |
Su L, Pang S J, Shan T F, Li X. 2017. Large-scale hatchery of the kelp Saccharina japonica: a case study experience at Lvshun in northern China. Journal of Applied Phycology, 29(6): 3 003-3 013.
DOI:10.1007/s10811-017-1154-y |
Sun X, Wu J, Wang G C, Kang Y N, Ooi H S, Shen T T, Wang F J, Yang R, Xu N J, Zhao X D. 2018. Genomic analyses of unique carbohydrate and phytohormone metabolism in the macroalga Gracilariopsis lemaneiformis (Rhodophyta). BMC Plant Biology, 18(1): 94.
DOI:10.1186/s12870-018-1309-2 |
Tai S H, Fang T C. 1977. The chromosomes of Laminaria japonica Aresch. Acta Genetica Sinica, 4(4): 325-329.
(in Chinese with English abstract) |
Tseng C K, Sun K Y, Wu C Y. 1955. On the cultivation of Haitai (Laminaria japonica Aresch.) by summering young sporophytes at low temperature. Acta Botanica Sinica, 4(3): 255-264.
(in Chinese with English abstract) |
Tseng C K, Wu C Y, Ren K Z. 1962. The influence of temperature on the growth and development of the Haidai (Laminaria japonica) gametophytes. Oceanologia et Limnologia Sinica, 4(1-2): 22-28.
(in Chinese with English abstract) |
Tseng C K, Wu C Y, Sun K Y. 1957. The effect of temperature on the growth and development of Haitai (Laminaria japonica Aresch.). Acta Botanica Sinica, 6(2): 103-130.
(in Chinese with English abstract) |
Tseng C K. 1981. Commercial cultivation. In: Lobban C S, Wynne M J eds. The Biology of Seaweeds. Blackwell Scientific Publications, Oxford. p.680-725.
|
Wang G L, Tan X L, Shen J L, Li J, Zhang L, Sun J W, Wang B, Weng M L, Liu T. 2011. Development of EST-SSR primers and their practicability test for Laminaria. Acta Oceanologica Sinica, 30(3): 112-117.
DOI:10.1007/s13131-011-0125-4 |
Wang J F, Jiang P, Cui Y L, Deng X Y, Li F C, Liu J G, Qin S. 2010. Genetic transformation in Kappaphycus alvarezii using micro-particle bombardment: a potential strategy for germplasm improvement. Aquaculture International, 18(6): 1 027-1 034.
DOI:10.1007/s10499-010-9320-0 |
Wang W J, Wang F J, Sun X T, Liu F L, Liang Z R. 2013a. Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae). Planta, 237(4): 1 123-1 133.
DOI:10.1007/s00425-012-1831-7 |
Wang X L, Chen Z H, Li Q Y, Zhang J, Liu S, Duan D L. 2018. High-density SNP-based QTL mapping and candidate gene screening for yield-related blade length and width in Saccharina japonica (Laminariales, Phaeophyta). Scientific Reports, 8(1): 13 591.
DOI:10.1038/s41598-018-32015-y |
Wang X L, Shao Z R, Fu W D, Yao J T, Hu Q P, Duan D L. 2013b. Chloroplast genome of one brown seaweed, Saccharina japonica (Laminariales, Phaeophyta): its structural features and phylogenetic analyses with other photosynthetic plastids. Marine Genomics, 10: 1-9.
DOI:10.1016/j.margen.2012.12.002 |
Wang X L, Yang Y X, Cong Y Z, Duan D L. 2004. DNA fingerprinting of selected Laminaria (Phaeophyta) gametophytes by RAPD markers. Aquaculture, 238(1-4): 143-153.
DOI:10.1016/j.aquaculture.2004.05.007 |
Wattier R, Maggs C A. 2001. Intraspecific variation in seaweeds: the application of new tools and approaches. Advances in Botanical Research, 35: 171-212.
DOI:10.1016/S0065-2296(01)35007-3 |
Wu C H, Jiang P, Guo Y, Liu J G, Zhao J, Fu H H. 2018. Isolation and characterization of Ulva prolifera actin1 gene and function verification of the 5' flanking region as a strong promoter. Bioengineered, 9(1): 124-133.
DOI:10.1080/21655979.2017.1325041 |
Wu C Y, Lin G H. 1987. Progress in the genetics and breeding of economic seaweeds in China. Hydrobiologia, 151-152: 57-61.
DOI:10.1007/BF00046105 |
Xu Y B. 2010. Molecular Plant Breeding. CABI, Cambridge, USA.
|
Xuan J S, Feng Y B, Weng M L, Zhao G, Shi J F, Yao J T, Wang X L, Guo B T, Qiao L X, Duan D L, Wang B. 2012. Expressed sequence tag analysis and cloning of trehalose-6-phosphate synthase gene from marine alga Laminaria japonica (Phaeophyta). Acta Oceanologica Sinica, 31(6): 139-148.
DOI:10.1007/s13131-012-0260-6 |
Yabu H, Yasui H. 1991. Chromosome number in four species of Laminaria (Phaeophyta). Japanese Journal of Phycology, 39(2): 185-187.
|
Yabu H. 1973. Alternation of chromosomes in the life history of Laminaria japonica Aresch. Bulletin of the Faculty of Fisheries Hokkaido University, 23(4): 171-176.
|
Yang G P, Sun Y, Shi Y Y, Zhang L, Guo S S, Li B J, Li X J, Li Z L, Cong Y Z, Zhao Y S, Wang W Q. 2009. Construction and characterization of a tentative amplified fragment length polymorphism-simple sequence repeat linkage map of Laminaria (Laminariales, Phaeophyta). Journal of Phycology, 45(4): 873-878.
DOI:10.1111/j.1529-8817.2009.00720.x |
Yang Q F, Liu L, Liu Y, Zhou Z G. 2017. Telomeric localization of the Arabidopsis-type heptamer repeat, (TTTAGGG)n, at the chromosome ends in Saccharina japonica (Phaeophyta). Journal of Phycology, 53(1): 235-240.
DOI:10.1111/jpy.12497 |
Yang X Q, Li L, Wang X L, Yao J T, Duan D L. 2020. Non-Coding RNAs participate in the regulation of CRY-DASH in the growth and early development of Saccharina japonica (Laminariales, Phaeophyceae). International Journal of Molecular Sciences, 21(1): 309.
DOI:10.3390/ijms21010309 |
Ye N H, Zhang X W, Miao M, Fan X, Zheng Y, Xu D, Wang J F, Zhou L, Wang D S, Gao Y, Wang Y T, Shi W Y, Ji P F, Li D M, Guan Z, Shao C W, Zhuang Z M, Gao Z Q, Qi J, Zhao F Q. 2015. Saccharina genomes provide novel insight into kelp biology. Nature Communications, 6(1): 6 986.
DOI:10.1038/ncomms7986 |
Ye R, Yu Z, Shi W W, Gao H J, Bi Y H, Zhou Z G. 2014. Characterization of α-type carbonic anhydrase (CA) gene and subcellular localization of α-CA in the gametophytes of Saccharina japonica. Journal of Applied Phycology, 26(2): 881-890.
DOI:10.1007/s10811-013-0221-2 |
Yoon H S, Lee J Y, Boo S M, Bhattacharya D. 2001. Phylogeny of Alariaceae, Laminariaceae, and Lessoniaceae (Phaeophyceae) based on plastid-encoded RuBisCo spacer and nuclear-encoded ITS sequence comparisons. Molecular Phylogenetics and Evolution, 21(2): 231-243.
DOI:10.1006/mpev.2001.1009 |
Yotsukura N, Kawai T, Motomura T, Ichimura T. 2001. Random amplified polymorphic DNA markers for three Japanese Laminarian species. Fisheries Science, 67(5): 857-862.
DOI:10.1046/j.1444-2906.2001.00333.x |
Yotsukura N, Maeda T, Abe T, Nakaoka M, Kawai T. 2016. Genetic differences among varieties of Saccharina japonica in northern Japan as determined by AFLP and SSR analyses. Journal of Applied Phycology, 28(5): 3 043-3 055.
DOI:10.1007/s10811-016-0807-6 |
Yotsukura N, Shimizu T, Katayama T, Druehl L D. 2010. Mitochondrial DNA sequence variation of four Saccharina species (Laminariales, Phaeophyceae) growing in Japan. Journal of Applied Phycology, 22(3): 243-251.
DOI:10.1007/s10811-009-9452-7 |
Zhang J, Li W, Qu J Q, Wang X M, Liu C, Liu T. 2015a. Development and characterization of microsatellite markers from an enriched genomic library of Saccharina japonica. Journal of Applied Phycology, 27(1): 479-487.
DOI:10.1007/s10811-014-0301-y |
Zhang J, Liu T, Bian D P, Zhang L, Li X B, Liu D T, Liu C, Cui J J, Xiao L Y. 2016a. Breeding and genetic stability evaluation of the new Saccharina variety "Ailunwan" with high yield. Journal of Applied Phycology, 28(6): 3 413-3 421.
DOI:10.1007/s10811-016-0810-y |
Zhang J, Liu T, Feng R F, Liu C, Chi S. 2015b. Genetic map construction and quantitative trait locus (QTL) detection of six economic traits using an F2 population of the hybrid from Saccharina longissima and Saccharina japonica. PLoS One, 10(5): e0128558.
DOI:10.1371/journal.pone.0128588 |
Zhang J, Liu T, Feng R F, Liu C, Jin Y M, Jin Z H, Song H Z. 2018a. Breeding of the new Saccharina variety "Sanhai" with high yield. Aquaculture, 485: 59-65.
DOI:10.1016/j.aquaculture.2017.11.015 |
Zhang J, Liu T, Rui F P. 2018b. Development of EST-SSR markers derived from transcriptome of Saccharina japonica and their application in genetic diversity analysis. Journal of Applied Phycology, 30(3): 2 101-2 109.
DOI:10.1007/s10811-017-1354-5 |
Zhang J, Liu Y, Yu D, Song H Z, Cui J J, Liu T. 2011. Study on high-temperature-resistant and high-yield Laminaria variety "Rongfu". Journal of Applied Phycology, 23(2): 165-171.
DOI:10.1007/s10811-011-9650-y |
Zhang J, Wang X L, Yao J T, Li Q Y, Liu F L, Yotsukura N, Krupnova T N, Duan D L. 2017. Effect of domestication on the genetic diversity and structure of Saccharina japonica populations in China. Scientific Reports, 7(1): 42158.
DOI:10.1038/srep42158 |
Zhang J, Wang X L, Yao J T, Yotsukura N, Duan D L. 2019a. Screening of polymorphic microsatellites and their application for Saccharina angustata and Saccharina longissima population genetic analysis. Journal of Applied Phycology, 31(5): 3 295-3 301.
DOI:10.1007/s10811-019-01798-6 |
Zhang J, Yao J T, Hu Z M, Jueterbock A, Yotsukura N, Krupnova T N, Nagasato C, Duan D L. 2019b. Phylogeographic diversification and postglacial range dynamics shed light on the conservation of the kelp Saccharina japonica. Evolutionary Applications, 12(4): 791-803.
DOI:10.1111/eva.12756 |
Zhang J, Yao J T, Sun Z M, Fu G, Galanin D A, Nagasato C, Motomura T, Hu Z M, Duan D L. 2015c. Phylogeographic data revealed shallow genetic structure in the kelp Saccharina japonica (Laminariales, Phaeophyta). BMC Evolutionary Biology, 15(1): 237.
DOI:10.1186/s12862-015-0517-8 |
Zhang L, Cui C J, Li Y, Wu H, Li X J. 2018c. A genome screen for the development of sex-specific DNA markers in Saccharina japonica. Journal of Applied Phycology, 30(2): 1 239-1 246.
DOI:10.1007/s10811-017-1295-z |
Zhang L, Li J K, Wu H, Li Y X. 2019c. Isolation and expression analysis of a candidate gametophyte sex determination gene (SJHMG) of kelp (Saccharina japonica). Journal of Phycology, 55(2): 343-351.
DOI:10.1111/jpy.12821 |
Zhang L, Peng J, Li X J, Cui C J, Sun J, Yang G P. 2016b. Characterization of genome-wide microsatellites of Saccharina japonica based on a preliminary assembly of Illumina sequencing reads. Journal of Ocean University of China, 15(3): 523-532.
DOI:10.1007/s11802-016-2945-5 |
Zhang L, Peng J, Li X J, Liu Y L, Cui C J, Wu H, Wu R N, Tian P P, Li Y. 2014. Development of 27 trinucleotide microsatellite markers for Saccharina japonica using next generation sequencing technology. Conservation Genetics Resource, 6(2): 341-344.
DOI:10.1007/s12686-013-0089-0 |
Zhang L, Wang X M, Liu T, Wang G L, Chi S, Liu C, Wang H Y. 2015d. Complete plastid genome sequence of the brown alga Undaria pinnatifida. PLoS One, 10(10): e0139366.
DOI:10.1371/journal.pone.0139366 |
Zhang L, Wang X M, Liu T, Wang H Y, Wang G L, Chi S, Liu C. 2015e. Complete plastid genome of the brown alga Costaria costata (Laminariales, Phaeophyceae). PLoS One, 10(10): e0140144.
DOI:10.1371/journal.pone.0140144 |
Zhang N, Zhang L N, Tao Y, Guo L, Sun J, Li X, Zhao N, Peng J, Li X J, Zeng L, Chen J S, Yang G P. 2015f. Construction of a high density SNP linkage map of kelp (Saccharina japonica) by sequencing Taq Ⅰ site associated DNA and mapping of a sex determining locus. BMC Genomics, 16(1): 189.
DOI:10.1186/s12864-015-1371-1 |
Zhang P Y, Shao Z R, Jin W H, Duan D L. 2016c. Comparative characterization of two GDP-mannose dehydrogenase genes from Saccharina japonica (Laminariales, Phaeophyceae). BMC Plant Biology, 16(1): 62.
DOI:10.1186/s12870-016-0750-3 |
Zhang P Y, Shao Z R, Li L, Liu S, Yao J L, Duan D L. 2018d. Molecular characterisation and biochemical properties ofphosphomannomutase/phosphoglucomutase (PMM/ PGM) in the brown seaweed Saccharina japonica. Journal of Applied Phycology, 30(4): 2 687-2 696.
DOI:10.1007/s10811-018-1460-z |
Zhang Q S, Qu S C, Cong Y Z, Luo S J, Tang X X. 2008a. High throughput culture and gametogenesis induction of Laminaria japonica gametophyte clones. Journal of Applied Phycology, 20(2): 205-211.
DOI:10.1007/s10811-007-9220-5 |
Zhang Q S, Tang X X, Cong Y Z, Qu S C, Luo S J, Yang G P. 2007. Breeding of an elite Laminaria variety 90-1 through inter-specific gametophyte crossing. Journal of Applied Phycology, 19(4): 303-311.
DOI:10.1007/s10811-006-9137-4 |
Zhang Y C, Jiang P, Gao J T, Liao J M, Sun S J, Shen Z L, Qin S. 2008b. Recombinant expression of rt-PA gene (encoding reteplase) in gametophytes of the seaweed Laminaria japonica (Laminariales, Phaeophyta). Science in China Series C: Life Sciences, 51(12): 1 116-1 120.
DOI:10.1007/s11427-008-0143-4 |
Zhou L R, Dai J X, Shen S D. 2004. An improved chromosome preparation from male gametophyte of Laminaria japonica (Heterokontophyta). Hydrobiologia, 512(1-3): 141-144.
DOI:10.1023/B:HYDR.0000020319.36532.eb |
Zhou W, Hu Y Y, Sui Z H, Fu F, Wang J G, Chang L P, Guo W H, Li B B. 2013. Genome survey sequencing and genetic background characterization of Gracilariopsis lemaneiformis (Rhodophyta) based on next-generation sequencing. PLoS One, 8(7): e69909.
DOI:10.1371/journal.pone.0069909 |
 2020, Vol. 38
2020, Vol. 38