Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Shasha, ZHENG Yingqiu, CHEN Muyan, B. STOREY Kenneth
- Ultrastructural variation and key ER chaperones response induced by heat stress in intestinal cells of sea cucumber Apostichopus japonicus
- Journal of Oceanology and Limnology, 39(1): 317-328
- http://dx.doi.org/10.1007/s00343-020-9265-8
Article History
- Received Nov. 4, 2019
- accepted in principle Feb. 19, 2020
- accepted for publication Mar. 14, 2020
2 Institute of Biochemistry, Carleton University, Ottawa ON K1S 5B6, Canada
In all eukaryotic cells, the endoplasmic reticulum (ER) is the central site for synthesis and folding of membrane and secretory proteins, and also participates in several important cellular functions including Ca2+ storage and cell signaling (Bánhegyi et al., 2007; Fonseca et al., 2011). The lumen of the ER provides an oxidative environment that is specialized for the formation of disulfide bonds and proper protein folding for secretion or transportation. It has been proposed that 30% newly synthesized proteins are rapidly degraded, possibly as a result of improper protein folding (Schubert et al., 2000; Yewdell and Nicchitta, 2006). Thus, the situation becomes even more critical if there are any perturbations of these processes (Lai et al., 2007). The accumulation of unfolded or misfolded proteins in the ER induces the unfolded protein response (UPR), which is the major protective and compensatory mechanism used by the ER to adapt to changing environmental or stress conditions and, thereby, re-establish normal ER function (Schröder, 2006). These adaptive mechanisms of the UPR involve transcriptional programs that promote ER-associated protein degradation to remove misfolded proteins, or induce up-regulation of genes such as those coding for resident chaperones and folding enzymes (Ma and Hendershot, 2004) that enhance the protein folding capacity of the ER (Xu et al., 2005). Autophagy, as an intracellular degradative system, plays important roles in regulating protein homeostasis, and is essential for survival when cells are faced with various physiological or pathological stresses (Levine and Klionsky, 2004; Yorimitsu and Klionsky, 2005). During autophagy, double membrane vesicles termed autophagosomes are formed, which sequester damaged cytosolic proteins or organelles as cargoes and then fuse with vacuoles or lysosomes (Yorimitsu et al., 2006). Mounting evidence shows that the autophagic response can be induced by ER stress in eukaryotes (ranging from yeast to mammals) and is essential for normal cellular development and differentiation (McCullough et al., 2001; Back et al., 2005; Iwata et al., 2005). In addition to act in the repair of unfolded or misfolded proteins that accumulate under ER stress, ER chaperones are also primary sensors of ER stress in response to hypoxia, virus infection, starvation and heat shock in many animals including sea cucumbers (Ma and Hendershot, 2004; Wang et al., 2015). Previous studies have shown that these chaperones such as glucose regulated protein 94 (GRP94), endoplasmic reticulum resident protein 29-like (ERP29) and protein disulfideisomerase A6-like (PDIA6) act to fold new proteins efficiently and refold stress-damaged proteins into their proper conformation (Hendrick and Hartl, 1995). However, very few studies have focused on the possible role of the UPR under adverse conditions in marine invertebrates (Chen et al., 2019).
The sea cucumber Apostichopus japonicus is a common species with high economic and medicinal values along the coasts of eastern Asia (Liao, 1997; Saito et al., 2002). As a typical temperate species, sea cucumbers are highly sensitive to changing variables of the marine environment (e.g. temperature, pH, salinity, etc.) (Asha and Muthiah, 2005; Wang et al., 2008). For example, temperature, as one of the most important abiotic factors, significantly affects the normal growth and metabolism of A. japonicus (Dong et al., 2007; Dong and Dong, 2008; Shao et al., 2015). In recent years, rapid temperature increasing and extreme high temperatures in the summer months has led to mass mortality of A. japonicus along the northern coasts of China, resulting in a dramatic resource decline and enormous economic loss, particularly for the aquaculture industry (Xu et al., 2017). Therefore, it is very important to understand the underlying molecular responses to heat stress in A. japonicus and identify regulatory mechanisms that can act to counteract effects of heat stress. The intestine of A. japonicus, the major tissue involved in food digestion and absorption of nutrients, has been reported to be highly sensitive to environmental change and to be a very important candidate organ for studies of the molecular responses to stress (Liao, 1997; Xu et al., 2016; Wang et al., 2018; Xu et al., 2018). To date, research on the molecular responses to heat stress in A. japonicus has mainly focused on expression of specific genes/proteins such as the heat shock protein (HSP) family and on epigenetic modifications such as microRNAs at an organismal level (Zhao et al., 2011; Chen et al., 2013; Xu et al., 2014; Wang et al., 2016; Chen et al., 2018; Gao et al., 2019). Due to the difficulty and immaturity of sea cucumber intestinal cell isolation and culture techniques, few studies have been conducted for endoplasmic reticulum stress response at the cellular and molecular levels (Wang et al., 2018), which will allow us to better understand the cytoprotective response to acute heat stress in sea cucumber intestinal cells. In the present study, we isolated sea cucumber intestinal cells by collagenase digestion and subjected them to heat stress. Ultrastructural changes in the intestinal cells were observed and changes in the expression levels of two key chaperones involved in ER stress were detected. We hope our current study will provide a candidate cytoprotective mechanism acting to counteract effects of heat stress in this marine invertebrate and help for heat-resistant variety selection of this valued economic species.
2 MATERIAL AND METHOD 2.1 Animal protocolsTwenty-one adult sea cucumbers A. japonicus (100±8 g) were collected from the coast of Qingdao (Jiaozhou Bay of the Yellow Sea, China) in May when seawater temperature was about 15 ℃. The animals were acclimated in seawater aquarium system at 15 ℃ for one week at the Ocean University of China and fed with an artificial food compound (including kelp, scallop skirt, brown alga (Sargassum thunbergii), gulfweed, Ulva and yeast) every morning. Subsequently, intestine samples from the mid-section of sea cucumbers were isolated and digested to release cells for experimental analysis. All animal procedures and experimental protocols were in compliance with the guidelines of the Experimental Animal Ethics Committee of Ocean University of China.
2.2 Cell culture and heat treatmentThe cell culture and heat treatment were performed as previously described by Wang et al. (2018). Briefly, isolated intestines of A. japonicus from 12 individuals were treated with 0.125% collagenase IA (c9891, Sigma, USA) at 22 ℃ for 2–3 h as described previously (Odintsova et al., 2005). Cell suspension was obtained after centrifugation at the speed of 300×g for 2 min at 4 ℃ and filtered through cell strainer with a 70-μm mesh size. The eluted cell suspension was washed three times for 10 min each time in the artificial seawater suspending (CMFSS) dissolved with neomycin (10 mg/mL, N8090, Solarbio, Beijing, China) and amphotericin B (2.5 mg/ mL, A8250, Solarbio, Beijing, China), followed by centrifugation for 10 min at 900×g. Then the cell pellets was resuspended in a modified Leibovitz's L-15 medium (GNAM1300, Genom, Hangzhou, China) in 1.5-mL polyethylene tubes (Bello et al., 2015), and stabilized at 15 ℃ for half an hour for following experiments. Then all cells were then randomly divided into either control or experimental groups. The cells in the experimental group were heated at 25 ℃ for 4 h and were sampled at the same time with control group (15 ℃).
2.3 Transmission electron microscopySamples used for transmission electron microscopy (TEM) were processed using standard techniques (Ullman et al., 2008). Briefly, the intestinal cells (from 15 ℃ and 25 ℃ groups) were centrifuged and fixed with 2.0% glutaraldehyde in seawater (pH 7.4, 1.1 Osm) overnight at 4 ℃. After fixation, samples were placed in 1% osmium tetroxide with seawater for 2 h at room temperature and rapidly washed twice with sterile distilled water. Then, the samples were dehydrated in a graded series of ethyl alcohol and embedded in a mixture of Araldite and Epon 812. Ultrathin sections of 70–80 nm were cut with a Reichert-Jung Ultracut E ultramicrotome (Reichert, Austria) and placed on the Formvar coated slot copper grids. Sections were then counterstained with uranyl acetate followed by aqueous lead citrate and examined with a Jeol JEM 1200 transmission electron microscope (Japan). Digital images were acquired with an AMT XR-60 CCD Digital Camera System.
2.4 RNA isolation, cDNA synthesis, qRT-PCR and gene structure analysisRNeasy mini kit (74104, Qiagen, Germany) with RNase-free DNase-treatment (79254, Qiagen, Germany) was applied to extract total RNA from isolated intestinal cells following the manufacturer's instructions. For qRT-PCR analysis of intestinal cells in experiment group (25 ℃) and control group (15 ℃), first-strand cDNA was synthesized using PrimerScript™ RT reagent Kit (RR047A, TaKaRa, China) as the qPCR template, and the expression levels were measured in StepOnePlus (ABI Inc., USA) using SYBR® Premix Ex TaqTM (RR420, TaKaRa, Japan). Six biological replicates were detected for each group and cells from two individual intestines pooled as a biological replicate. Each sample was run in triplicate. The specific primers for AjERP29 (PIK36082.1) and AjPDIA6 (PIK49878.1) were listed in Table 1. β-tubulin (TUBB, PIK51093) and β-Actin (ACTB, PIK61412.1) were selected as the internal controls, according to previous studies (Zhao et al., 2014; Ono et al., 2018). Melting-curve analysis of the amplification products was performed at the end of the PCR reaction to confirm specificity.
The complete sequences of AjERP29 and AjPDIA6 were obtained from the A. japonicus genome reported by Zhang et al. (2017), which is publicly available from GenBank with accession number GCA_002754855.1. The promoter region sequence was predicted by Promoter 2.0 Prediction Server (http://www.cbs.dtu.dk/services/Promoter/). The transcription factor binding sites in the 5′-flanking region were predicted using the AiBaba 2.1 programs (http://gene-regulation.com/pub/programs/alibaba2/index.html). Multiple sequence alignments and phylogenetic-tree construction applying the neighbor joining (NJ) method were conducted using the ClustalW program and MEGA 6.0 software.
2.5 Preparation of cell protein extractsThree biological replicates (cells from three individuals pooled as a biological replicate) were used for total protein extraction in treatment (25 ℃) and control group (15 ℃), respectively. Cell lysis buffer (phenylmethylsulfonyl fluoride added) (P0013, Beyotime, Beijing, China) was mixed gently with intestinal cells on ice after centrifuging at the speed of 100×g for 10 min. Then centrifuging (14 000×g, 5 min, 4 ℃) to collect supernatant. Enhanced BCA protein assay kit (P0010S, Beyotime, Beijing, China) was used to measure soluble protein concentration in SpectraMax Plus (Molecular Devices, USA) system. The qualified protein was stored at -80 ℃ for use.
2.6 Protein Expression level of AjERP29 and AjPDIA6The Western blot was applied to analyze the protein expression levels following the protocol as previously described (Chen et al., 2016). In brief, the protein samples (15–25 μg) were mixed with 4×Laemmli buffer (1610747.0, Beyotime, Beijing, China) and heated at 95 ℃ for 5 min. For sodium dodecyl sulfate polyacrylamide gels electrophoresis (SDS-PAGE), equal protein amounts were loaded into adjacent lanes on 12% gels and run for 2 h at 120V. Proteins were then transferred to polyvinylidene fluoride membrane (PVDF) (IPVH00010, Bedford, MA, USA) for 30 or 45 min at 280 mA following a wet transfer procedure (BioRad, USA). The membranes were blocked with 5% non-fat powdered milk in TBST (Tris Buffered Saline with Tween 20) for 2 h at room temperature and then washed five times for 10 min each on a gently shaking platform. Membranes were then incubated with one of the following primary antibodies: rabbit polyclonal antibody against AjERP29 (0.464 mg/mL, diluted 1:450 for cells in TBS with 5% non-fat powdered milk and 0.05% Tween-20, prepared by Genscipt, Nanjing, China) and rabbit monoclonal antibody against AjPDIA6 (0.364 mg/mL, diluted 1꞉330 in TBS with 5% non-fat powdered milk and 0.05% Tween-20, prepared by Genscipt, Nanjing, China) overnight at 4 ℃ with gentle shaking. The membranes were then washed with TBST five times for 10 min each and incubated with goat anti-rabbit IgG conjugated to horseradish peroxidase (1:10 000, 2578S, CST, Danvers, MA, USA) for 1 h at room temperature. β-Tubulin was chosen as the internal protein standard and incubated with rabbit polyclonal β-Tubulin antibody (1꞉1 000, 2146S, CST, Danvers, MA, USA) and goat anti-rabbit lgG, HRP-linked antibody (1꞉10 000, 7074S, CST, Danvers, MA, USA). The membranes were then washed with TBST five times for 10 min each and bound antibodies were detected by an ECL detection reagent (P0018, Beyotime, Beijing, China) according to the manufacturer's instructions, followed by semiquantification analysis using the Image-Pro Plus 6.0 software (Media Cybernetics Inc., USA). Individual blots were repeated 3 times
2.7 Statistical analysisRelative expression levels of AjERP29 and AjPDIA6 at the level of transcription were evaluated by 2-ΔΔCt method. Values were normalized against the β-Tubulin and β-Actin transcripts, respectively, and then mean values were calculated. The results were statistically analyzed using the Student's t-test with P < 0.05 accepted as statistically significant (unless otherwise stated). All data are presented as mean±SE (n=6 separate isolations of intestinal cells).
Relative protein abundance of AjERP29 and AjPDIA6 was normalized by β-Tubulin (Wang et al., 2018; Wang et al., 2019) and analyzed using the Student's t-test with P < 0.05 accepted as statistically significant. Results are presented as mean±SE (n=3 independent protein isolations from different animals).
3 RESULT AND DISCUSSION 3.1 Homology and phylogenetic analyses of AjERP29 and AjPDIA6Alignment of the deduced amino acid sequences of AjERP29 from A. japonicus with the amino acid sequences of ERP29 from other species showed the highest homology with the following marine invertebrates: Acanthaster planci (starfish, 51% identity), Lingula anatina (lingula, 48% identity), Branchiostoma belcheri (lancelet, 47% identity) and Crassostrea gigas (oyster, 47% identity) (Fig. 1a). A phylogenetic tree based on the NJ method was constructed by analyzing the predicted amino acid sequences of ERP29 from different species (Fig. 1b). The predicted AjERP29 sequence from A. japonicus clustered into a clade with another echinoderm species (starfish), and distinct from other invertebrate clades (Fig. 1b).
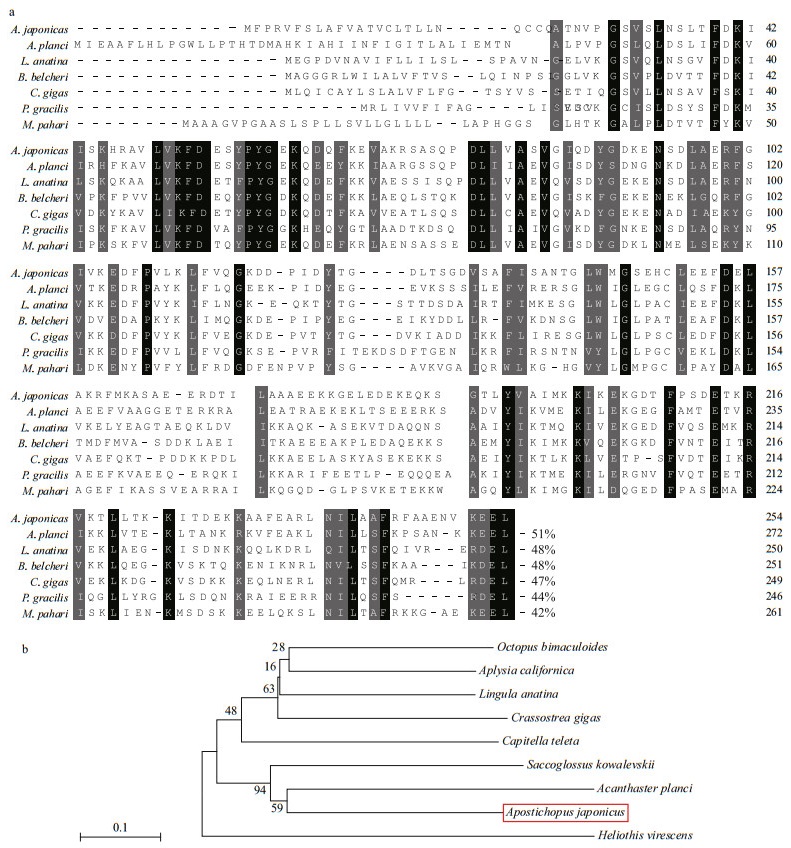
|
| Fig.1 Homology and phylogenetic analyses of the deduced amino acid sequence of AjERP29 compared with ERP29 from other invertebrate species a. comparison of the amino acid sequence of sea cucumber ERP29 with the protein from other species. Identical and similar amino acid residues are indicated by black and grey highlights, respectively. The percent identity of each amino acid sequence to AjERP29 is shown at the end of each sequence; b. sequences were acquired from GenBank: Lingula anatine (lingula), XP_013403746.1; Crassostrea gigas (Pacific oyster), XP_011428502.1; Heliothis virescens (tobacco budworm), PCG78617.1; Capitella teleta (a polychaete annelid worm), ELT98573.1; Saccoglossus kowalevskii (acorn worm), XP_002736824.1; Acanthaster planci (crown-of-thorns sea star), XP_022108445.1; Aplysia californica (California sea hare), XP_005102188.1; Octopus bimaculoides (two-spot octopus), XP_014782649.1. Mus pahari (gairdner's shrew-mouse), XM_021187221.2; Pseudomyrmex gracilis (leaf-cutting ants), XM_020436368. 1; Branchiostoma belcheri (lancelet amphioxus), XM_019760885.1. |
The deduced amino acid sequence of AjPDIA6 from A. japonicus showed the highest identities of 59%, 59%, and 58% with those of Strongylocentrotus purpuratus (sea urchin), Acanthaster planci (starfish), and Lingula anatina (lingula), respectively (Fig. 2a). A phylogenetic tree based on the NJ method was constructed by analyzing the predicted amino acid sequences of PDIA6 from different species (Fig. 2b). The amino acid sequence of AjPDIA6 was also clustered into one clade with other echinoderms (sea urchin and starfish) (Fig. 2b) and distincted from the vertebrate clade. The results for both proteins indicated that the evolution of ERP29 and PDIA6 are consistent with the known evolutionary position of echinoderm species. Furthermore, the high conservation of these two proteins across different animal phyla suggests that they may share a common important function across the animal kingdom.
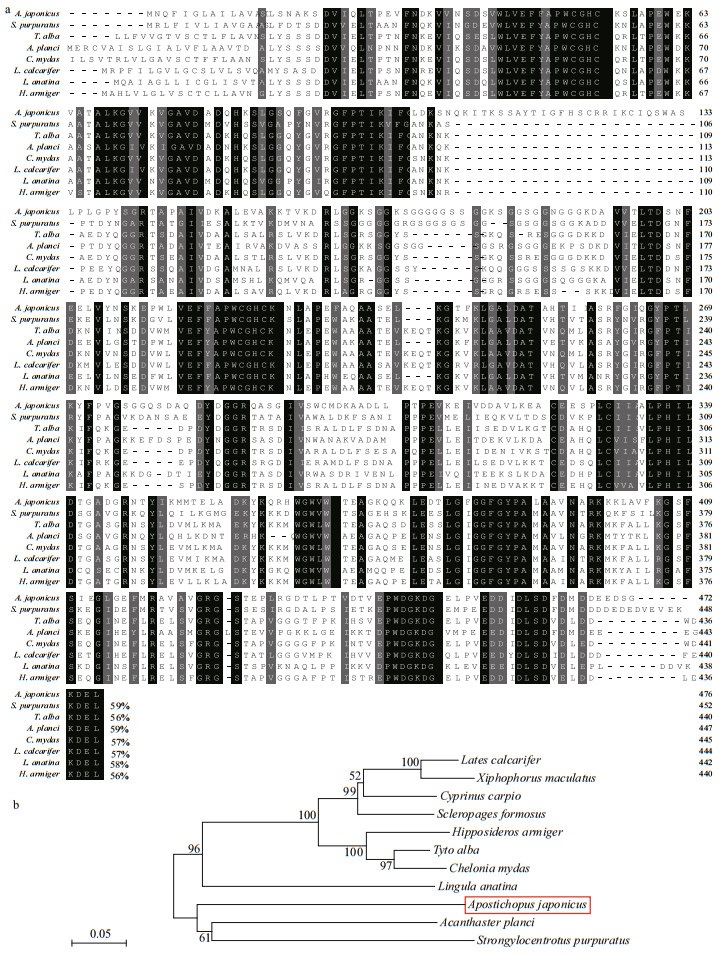
|
| Fig.2 Homology and phylogenetic analyses of the deduced amino acid sequence of AjPDIA6 with PDIA6 from other species a. comparison of the amino acid sequence of sea cucumber PDIA6 with PDIA6 from other species. Identical and similar amino acid residues are indicated by black and grey highlights, respectively. Percent identity of amino acid sequence to AjPDIA6 is shown at the end of each sequence; b. phylogenetic tree based on the sequences of PDIA6 from different species. The tree topology was evaluated by 1 000 bootstrap replications and numbers on each branch of the tree represent the bootstrap support value. PDIA6 sequences were acquired from GenBank: Tyto alba (barn owl), KFV46902.1; Scleropages formosus (Asian arowana), XP_018589931.1; Lingula anatine (lingula), XP_013413636.1; Cyprinus carpio (European carp), XP_018981098.1; Hipposideros armiger (great roundleaf bat), XP_019505574.1; Xiphophorus maculatus (platyfish), XP_005803193.1; Lates calcarifer (barramundi), XP_018550486.1; Strongylocentrotus purpuratus (purple sea urchin), XP_790496.3; Acanthaster planci (crown-of-thorns seastar), XP_022083302.1; Chelonia mydas (green sea turtle), XP_007053233.1. |
We also observed a four-residue ER-retention motif (KEEL for AjERP29 and KDEL for AjPDIA6) at the carboxyl-terminus of the translated sequence (Fig. 1a & b). These are typical characteristics of reticuloplasmins residing in the ER lumen (Scheel and Pelham, 1996), suggesting that both proteins are reticuloplasmins.
3.2 Ultrastructural observations of sea cucumber intestinal cells in response to heat stressObvious structural changes in intestine tissue under high temperature stress have been reported and studied previously in A. japonicus (Czechowski, 1996; DeSesso and Williams, 2008). Cell apoptosis after 192 h of heat stress and intensified ultrastructural damage with increasing time was also reported in A. japonicus (Xu et al., 2015). The present study provided the first ultrastructural analysis of isolated intestinal cells in A. japonicus after heat stress (Fig. 3a & b). The most obvious structural change in the intestinal cells under heat stress was the expansion of the rough endoplasmic reticulum (Fig. 3b, white arrowhead), which means that the diameter of the endoplasmic reticulum increased, but still kept its original structure and a mesh shape. It is interesting to note that we also observed numerous autophagosomes in the lumen of heat-treated cells (Fig. 3b, black arrowhead) in comparison with control conditions. The images also show that the numbers of mitochondria and intracellular vesicles (lysosomes, residual bodies) decreased in stressed cells compared with the control group, suggesting that metabolic capacity and endocytosis/exocytosis functions were weakened in heat-stressed cells. Previous studies have indicated that ER stress is a potent inducer of autophagy, which resulted either in enhancing cell survival or in committing the cell to non-apoptotic death (Yorimitsu et al., 2006; Senft and Ronai, 2015). Thus, we deduced that an activation of autophagy may be induced through ER stress in sea cucumber intestinal cells under heat stress. Noticeably, the direct link between ER stress and autophagy has been rarely reported so far, and a series of questions such as the signal transduction pathway connecting ER stress and the role of autophagy induced by ER stress, remain unknown (Kabir et al., 2018). Ultrastructural changes caused by heat shock have been observed in various organelles inside the cells of other species. In spermatid cells of rats, mitochondrial degeneration, dilatation of smooth endoplasmic reticulum (SER) and enlarged intercellular spaces were recorded after hyperthermia (Kanter et al., 2013). Concentric stacking of rough endoplasmic reticulum (RER) and swelling of mitochondrial saccules were also reported under heat stress in tobacco Nicotiana sylvestris (Kandasamy and Kristen, 1989).

|
| Fig.3 Changes in the ultrastructure of sea cucumber intestinal cells under heat stress a. intestinal cells cultured at 15 ℃ as the control; b. intestinal cells heated at 25 ℃ for 4 h. Under heat stress, autophagosomes (black arrowhead) are scattered in the cytoplasm and the expansion of rough endoplasmic reticulum (white arrowhead) is obvious (m: mitochondria; ly: lysosome; n: nucleus; nu: nucleolus; rb: residual body). Scale bar: 1 μm. |
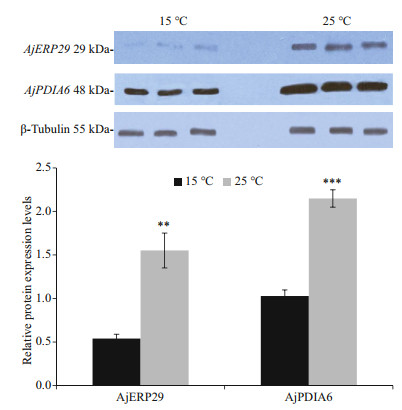
Promoters of AjERP29 and AjPDIA6 were predicted and analyzed. Interestingly, in our present study, C/EBPbeta binding site was observed in both of AjERP29 and AjPDIA6 promoter regions (Fig. 4) and it has been reported that the C/EBPbeta gene is induced in response to ER stress (Chen et al., 2004), which suggested that the variation of these two chaperones are potentially responsible for ER stress by binding with the activated C/EBPbeta gene. Growing evidence has suggested that reticuloplasmins might share common functions as a molecular chaperones during protein assembly and degradation and undergo up-regulated expression in response to cellular stresses such as improper protein folding or disrupted calcium homeostasis in the ER induced by biotic and abiotic factors (Kozutsumi et al., 1988; Harding et al., 2003; Ma and Hendershot, 2004; Schröder and Kaufman, 2005; Yin et al., 2017). The ER chaperones in these circumstances such as GRP94, ERP29 and PDI are primary sensors of ER stress and assist in repairing unfolded or misfolded proteins that accumulate in the ER under stress (Mkrtchian et al., 1998; Ma and Hendershot, 2002). In present study, we focused on the relative transcriptional and translational expression levels of EPR29 and PDIA6 in intestinal cells in response to heat stress. ERP29 has been shown to interact with and enhance the function of other ER chaperones, such as GRP94, GRP78, ERP72, and calnexin (Park et al., 2005), which suggested the important role of ERP29 in the regulation of cell survival and apoptosis. Protein disulfide isomerase A6-like (PDIA6) can also be considered as a key ER chaperone and is a multifunctional protein with a modular domain structure (Noiva and Lennarz, 1992). PDIA6 has been reported to participate in protein folding, assembly and post-translational modification by catalyzing native disulfide bond formation and rearrangement (Freedman et al., 1994; Tu and Weissman, 2004). Our present study showed that expression levels of AjERP29 mRNA transcripts increased significantly by 6.00 fold (P < 0.05) in intestinal cells in response to heat stress at 25 ℃ (Fig. 5) and that AjERP29 protein expression levels also rose significantly by 2.88 fold under heat stress, as compared with controls (P < 0.05) (Fig. 6). In addition, the transcriptional expression of AjPDIA6 was strongly increased by 3.66 fold at 25 ℃ (P < 0.05) (Fig. 5) and the protein expression levels of AjPDIA6 rose by 2.09 fold in response to heat stress, compared to controls (P < 0.05) (Fig. 6). Thus, we propose that AjERP29 and AjPDIA6 play important roles in the regulation of cell survival by alleviating ER stress caused by high temperatures in sea cucumbers. Previous studies have reported the overexpression of ERP29 and PDIA6 during heat stress associated with aestivation in sea cucumber intestinal tissue and coelomocytes (Zhang et al., 2013; Chen et al., 2016), which is supportive of our present study.
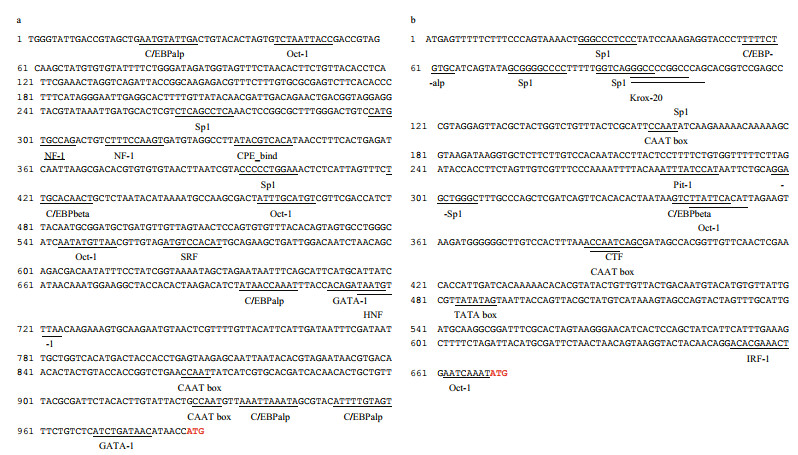
|
| Fig.4 The putative transcriptional factor binding sites in the promoter sequence of AjERP29 (a) and AjPDIA6 (b) genes The putative binding sites of the transcription factors are underlined. Initiation codon (ATG) is in red. |
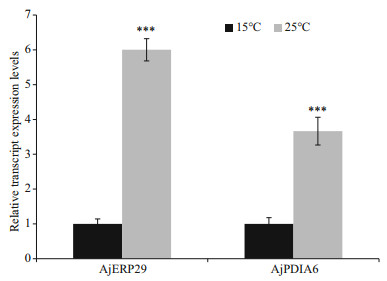
|
| Fig.5 Relative transcript expression levels of AjERP29 and AjPDIA6 in intestinal cells in control (15 ℃) and heat treatment (25 ℃) groups, respectively Values are mean±SE (n=6). *** indicates significant difference from the control, P < 0.001. TUBB and ACTB were used as the reference genes. |
|
| Fig.6 Relative protein expression levels of AjERP29 and AjPDIA6 in control group (15 ℃) and experiment group (25 ℃) using western blot Representative bands represent the immunoblot intensities and histograms show the standardized levels for 15 ℃ and 25 ℃, respectively. Values are means±SE (n=3). ***: indicates significant difference from the control, P < 0.001; ** P < 0.01. |
Heat stress may greatly influence the survival of sea cucumbers in summer. To our knowledge, this study provides the first demonstration of an endoplasmic reticulum stress response at the cellular and molecular levels in sea cucumber intestinal cells and allows us to better understand the potential cytoprotective mechanism in response to heat stress in sea cucumber, Which may fill the knowledge of thermal adaptive mechanism in sea cucumber and provide support for heat-resistant variety selection of this valued economic species. Further studies will also be needed to gain additional details about the regulatory pathway of the ER stress response in this species.
5 DATA AVAILABILITY STATEMENTAll data generated and/or analyzed during this study are included in this published article and its supplementary information files.
6 COMPETING INTERESTS STATEMENTThe authors declare no competing interests.
Asha P S, Muthiah P. 2005. Effects of temperature, salinity and pH on larval growth, survival and development of the sea cucumber Holothuria spinifera Theel. Aquaculture, 250(3-4): 823-829.
DOI:10.1016/j.aquaculture.2005.04.075 |
Back S H, Schröder M, Lee K, Zhang K Z, Kaufman R J. 2005. ER stress signaling by regulated splicing:IRE1/HAC1/XBP1. Methods, 35(4): 395-416.
DOI:10.1016/j.ymeth.2005.03.001 |
Bánhegyi G, Baumeister P, Benedetti A, Dong D Z, Fu Y, Lee A S, Li J Z, Mao C H, Margittai E, Ni M, Paschen W, Piccirella S, Senesi S, Sitia R, Wang M, Yang W. 2007. Endoplasmic reticulum stress. Annals of the New York Academy of Sciences, 1113(1): 58-71.
DOI:10.1196/annals.1391.007 |
Bello S A, Abreu-Irizarry R J, García-Arrarás J E. 2015.Primary cell cultures of regenerating holothurian tissues.In: Nelson C M ed. Tissue Morphogenesis. Humana Press, New York. p.283-297, https://doi.org/ 10.1007/978-1-4939-1164-6_19.
|
Chen C, Dudenhausen E E, Pan Y X, Zhong C, Kilberg M S. 2004. Human CCAAT/enhancer-binding protein β gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. Journal of Biological Chemistry, 279(27): 27948-27956.
DOI:10.1074/jbc.M313920200 |
Chen M Y, Li X K, Zhu A J, Storey K B, Sun L N, Gao T X, Wang T M. 2016. Understanding mechanism of sea cucumber Apostichopus japonicus aestivation:insights from TMT-based proteomic study. Comparative Biochemistry and Physiology Part D:Genomics and Proteomics, 19: 78-89.
DOI:10.1016/j.cbd.2016.06.005 |
Chen M Y, Wang S S, Li X K, Storey K B, Zhang X M. 2018. The potential contribution of miRNA-200-3p to the fatty acid metabolism by regulating AjEHHADH during aestivation in sea cucumber. PeerJ, 6: e5703.
DOI:10.7717/peerj.5703 |
Chen M Y, Zhang X M, Liu J N, Storey K B. 2013. Highthroughput sequencing reveals differential expression of miRNAs in intestine from sea cucumber during aestivation. PLoS One, 8(10): e76120.
DOI:10.1371/journal.pone.0076120 |
Chen Y H, Lian Y Y, He H H, Yuan K, Zhang C Z, Yu G H, He J G. 2019. Functional characterization of an ER-stress responding Crustin gene in Litopenaeus vannamei. Fish & Shellfish Immunology, 84: 541-550.
DOI:10.1016/j.fsi.2018.10.047 |
Czechowski J. 1996. Conventional radiography and ultrasonography in the diagnosis of small bowel obstruction and strangulation. Acta Radiologica, 37(2): 186-189.
DOI:10.1080/02841859609173442 |
DeSesso J M, Williams A L. 2008. Contrasting the gastrointestinal tracts of mammals:factors that influence absorption. Annual Reports in Medicinal Chemistry, 43: 353-371.
DOI:10.1016/S0065-7743(08)00021-3 |
Dong Y W, Dong S L. 2008. Induced thermotolerance and expression of heat shock protein 70 in sea cucumber Apostichopus japonicus. Fisheries Science, 74(3): 573-578.
DOI:10.1111/j.1444-2906.2008.01560.x |
Dong Y W, Ji T T, Dong S L. 2007. Stress responses to rapid temperature changes of the juvenile sea cucumber(Apostichopus japonicus Selenka). Journal of Ocean University of China, 6(3): 275-280.
DOI:10.1007/s11802-007-0275-3 |
Fonseca S G, Gromada J, Urano F. 2011. Endoplasmic reticulum stress and pancreatic β-cell death. Trends in Endocrinology & Metabolism, 22(7): 266-274.
DOI:10.1016/j.tem.2011.02.008 |
Freedman R B, Hirst T R, Tuite M F. 1994. Protein disulphide isomerase:building bridges in protein folding. Trends in Biochemical Sciences, 19(8): 331-336.
DOI:10.1016/0968-0004(94)90072-8 |
Gao L, Yuan Z H, Ma Z, Li Z, Yu S M, Li Y F, He C B. 2019. Genome-wide comparative analysis of the SHSP, HSP60/10 and HSP90 genes reveals differential heat stress responses in estivation of the sea cucumber Apostichopus japonicus. Aquaculture Research, 50(4): 1117-1130.
DOI:10.1111/are.13986 |
Harding H P, Zhang Y H, Zeng H, Novoa I, Lu P D, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl D F, Bell J C, Hettmann T, Leiden J M, Ron D. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular Cell, 11(3): 619-633.
DOI:10.1016/S1097-2765(03)00105-9 |
Hendrick J P, Hartl F U. 1995. The role of molecular chaperones in protein folding. FASEB Journal, 9(15): 1559-1569.
DOI:10.1096/fasebj.9.15.8529835 |
Iwata A, Christianson J C, Bucci M, Ellerby L M, Nukina N, Forno L S, Kopito R R. 2005. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proceedings of the National Academy of Sciences of the United States of America, 102(37): 13135-13140.
DOI:10.1073/pnas.0505801102 |
Kabir M F, Kim H R, Chae H J. 2018. Endoplasmic reticulum stress and autophagy. In: Català A ed. Stress and Autophagy, Endoplasmic Reticulum. IntechOpen, London, https://doi.org/ 10.5772/intechopen.81381.
|
Kandasamy M K, Kristen U. 1989. Ultrastructural responses of tobacco pollen tubes to heat shock. Protoplasma, 153(1-2): 104-110.
DOI:10.1007/BF01322470 |
Kanter M, Aktas C, Erboga M. 2013. Heat stress decreases testicular germ cell proliferation and increases apoptosis in short term:an immunohistochemical and ultrastructural study. Toxicology and Industrial Health, 29(2): 99-113.
DOI:10.1177/0748233711425082 |
Kozutsumi Y, Segal M, Normington K, Gething M J, Sambrook J. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucoseregulated proteins. Nature, 332(6163): 462-464.
DOI:10.1038/332462a0 |
Lai E, Teodoro T, Volchuk A. 2007. Endoplasmic reticulum stress:signaling the unfolded protein response. Physiology, 22(3): 193-201.
DOI:10.1152/physiol.00050.2006 |
Levine B, Klionsky D J. 2004. Development by self-digestion:molecular mechanisms and biological functions of autophagy. Developmental Cell, 6(4): 463-477.
DOI:10.1016/S1534-5807(04)00099-1 |
Liao Y L. 1997. Fauna Sinica: Phylum Echinodermata: Class Holothuroidea. Science Press, Beijing. p.148-150. (in Chinese)
|
Ma Y J, Hendershot L M. 2002. The mammalian endoplasmic reticulum as a sensor for cellular stress. Cell Stress & Chaperones, 7(2): 222-229.
DOI:10.1379/1466-1268(2002)007<0222:tmeraa>2.0.co;2 |
Ma Y J, Hendershot L M. 2004. ER chaperone functions during normal and stress conditions. Journal of Chemical Neuroanatomy, 28(1-2): 51-65.
DOI:10.1016/j.jchemneu.2003.08.007 |
McCullough K D, Martindale J L, Klotz L O, Aw T Y, Holbrook N J. 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Molecular and Cellular Biology, 21(4): 1249-1259.
DOI:10.1128/MCB.21.4.1249-1259.2001 |
Mkrtchian S, Fang C, Hellman U, Ingelman-Sundberg M A. 1998. A stress-inducible rat liver endoplasmic reticulum protein, ERp29. European Journal of Biochemistry, 251(1-2): 304-313.
DOI:10.1046/j.1432-1327.1998.2510304.x |
Noiva R, Lennarz W J. 1992. Protein disulfide isomerase. A multifunctional protein resident in the lumen of the endoplasmic reticulum. Journal of Biological Chemistry, 267(6): 3553-3556.
|
Odintsova N A, Dolmatov I Y, Mashanov V S. 2005. Regenerating holothurian tissues as a source of cells for long-term cell cultures. Marine Biology, 146(5): 915-921.
DOI:10.1007/s00227-004-1495-3 |
Ono K, Suzuki T A, Toyoshima Y, Suzuki T, Tsutsui S, Odaka K, Miyadai T, Nakamura O. 2018. SJL-1, a C-type lectin, acts as a surface defense molecule in Japanese sea cucumber, Apostichopus japonicus. Molecular Immunology, 97: 63-70.
DOI:10.1016/j.molimm.2018.03.009 |
Park S, You K H, Shong M, Goo T W, Yun E Y, Kang S W, Kwon O Y. 2005. Overexpression of ERP29 in the thyrocytes of FRTL-5 cells. Molecular Biology Reports, 32(1): 7-13.
DOI:10.1007/s11033-004-3069-3 |
Saito M, Kunisaki N, Urano N, Kimura S. 2002. Collagen as the major edible component of sea cucumber (Stichopus japonicus). Journal of Food Science, 67(4): 1319-1322.
DOI:10.1111/j.1365-2621.2002.tb10281.x |
Scheel A A, Pelham H R B. 1996. Purification and characterization of the human KDEL receptor. Biochemistry, 35(31): 10203-10209.
DOI:10.1021/bi960807x |
Schröder M, Kaufman R J. 2005. The mammalian unfolded protein response. Annual Review of Biochemistry, 74: 739-789.
DOI:10.1146/annurev.biochem.73.011303.074134 |
Schröder M. 2006. The unfolded protein response. Molecular Biotechnology, 34(2): 279-290.
DOI:10.1385/MB:34:2:279 |
Schubert U, Antón L C, Gibbs J, Norbury C C, Yewdell J W, Bennink J R. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature, 404(6779): 770-774.
DOI:10.1038/35008096 |
Senft D, Ronai Z A. 2015. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends in Biochemical Sciences, 40(3): 141-148.
DOI:10.1016/j.tibs.2015.01.002 |
Shao Y N, Li C H, Chen X C, Zhang P J, Li Y, Li T W, Jiang J B. 2015. Metabolomic responses of sea cucumber Apostichopus japonicus to thermal stresses. Aquaculture, 435: 390-397.
DOI:10.1016/j.aquaculture.2014.10.023 |
Tu B P, Weissman J S. 2004. Oxidative protein folding in eukaryotes. Journal of Cell Biology, 164(3): 341-346.
DOI:10.1083/jcb.200311055 |
Ullman E, Fan Y, Stawowczyk M, Chen H M, Yue Z, Zong W X. 2008. Autophagy promotes necrosis in apoptosisdeficient cells in response to ER stress. Cell Death & Differentiation, 15(2): 422-425.
DOI:10.1038/sj.cdd.4402234 |
Wang F Y, Yang H S, Gao F, Liu G B. 2008. Effects of acute temperature or salinity stress on the immune response in sea cucumber, Apostichopus japonicus. Comparative Biochemistry and Physiology Part A:Molecular & Integrative Physiology, 151(4): 491-498.
DOI:10.1016/j.cbpa.2008.06.024 |
Wang H H, Li C H, Wang Z H, Shao Y N, Lv Z M, Zhang W W. 2016. p44/42MAPK and p90RSK modulate thermal stressed physiology response in Apostichopus japonicus. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 196-197: 57-66.
DOI:10.1016/j.cbpb.2016.02.008 |
Wang H H, Shao Y N, Zhang W W, Li C H, Lv Z M, Jin C H. 2015. Molecular characterization of two novel molecular chaperones in bacterial-challenged Apostichopus japonicus. Gene, 570(1): 141-149.
DOI:10.1016/j.gene.2015.06.024 |
Wang S S, Chen M Y, Yin Y C, Storey K B. 2019. MiR-200-3p is potentially involved in cell cycle arrest by regulating cyclin A during aestivation in Apostichopus japonicus. Cells, 8(8): 843.
DOI:10.3390/cells8080843 |
Wang S S, Li X K, Chen M Y, Storey K B, Wang T M. 2018. A potential antiapoptotic regulation:the interaction of heat shock protein 70 and apoptosis-inducing factor mitochondrial 1 during heat stress and aestivation in sea cucumber. Journal of Experimental Zoology Part A:Ecological and Integrative Physiology, 329(3): 103-111.
DOI:10.1002/jez.2180 |
Xu C Y, Bailly-Maitre B, Reed J C. 2005. Endoplasmic reticulum stress:cell life and death decisions. The Journal of Clinical Investigation, 115(10): 2656-2664.
DOI:10.1172/JCI26373 |
Xu D X, Sun L N, Liu S L, Zhang L B, Yang H S. 2014. Polymorphisms of heat shock protein 90 (Hsp90) in the sea cucumber Apostichopus japonicus and their association with heat-resistance. Fish & Shellfish Immunology, 41(2): 428-436.
DOI:10.1016/j.fsi.2014.09.025 |
Xu D X, Sun L N, Liu S L, Zhang L B, Yang H S. 2015. Histological, ultrastructural and heat shock protein 70(HSP70) responses to heat stress in the sea cucumber Apostichopus japonicus. Fish & Shellfish Immunology, 45(2): 321-326.
DOI:10.1016/j.fsi.2015.04.015 |
Xu D X, Sun L N, Liu S L, Zhang L B, Yang H S. 2016. Understanding the heat shock response in the sea cucumber Apostichopus japonicus, using iTRAQ-based proteomics. International Journal of Molecular Sciences, 17(2): 150.
DOI:10.3390/ijms17020150 |
Xu D X, Zhou S, Yang H S. 2017. Carbohydrate and amino acids metabolic response to heat stress in the intestine of the sea cucumber Apostichopus japonicus. Aquaculture Research, 48: 5883-5891.
DOI:10.1111/are.13411 |
Xu K, Yu Q H, Zhang J S, Lv Z M, Fu W D, Wang T M. 2018. Cell loss by apoptosis is involved in the intestinal degeneration that occurs during aestivation in the sea cucumber Apostichopus japonicus. Comparative Biochemistry and Physiology Part B:Biochemistry and Molecular Biology, 216: 25-31.
DOI:10.1016/j.cbpb.2017.11.004 |
Yewdell J W, Nicchitta C V. 2006. The DRiP hypothesis decennial:support, controversy, refinement and extension. Trends in Immunology, 27(8): 368-373.
DOI:10.1016/j.it.2006.06.008 |
Yin H C, Zhao L L, Jiang X J, Li S Q, Huo H, Chen H Y. 2017. DEV induce autophagy via the endoplasmic reticulum stress related unfolded protein response. PLoS One, 12(12): e0189704.
DOI:10.1371/journal.pone.0189704 |
Yorimitsu T, Klionsky D J. 2005. Autophagy:molecular machinery for self-eating. Cell Death & Differentiation, 12 Suppl 2: 1542-1552.
DOI:10.1038/sj.cdd.4401765 |
Yorimitsu T, Nair U, Yang Z F, Klionsky D J. 2006. Endoplasmic reticulum stress triggers autophagy. Journal of Biological Chemistry, 281(40): 30299-30304.
DOI:10.1074/jbc.M607007200 |
Zhang P, Lu Y L, Li C H, Su X R, Wang Z H, Jin C H, Li Y, Li T W. 2013. Identification of differential expressed proteins and characterization their mRNA expression in thermally stressed Apostichopus japonicus. Comparative Biochemistry and Physiology Part D:Genomics and Proteomics, 8(3): 194-200.
DOI:10.1016/j.cbd.2013.05.001 |
Zhang X j, Sun L N, Yuan J B, Sun Y M, Gao Y, Zhang L B, Li S H, Dai H, Hamel J F, Liu C Z, Yu Y, Liu S L, Lin W C, Guo K M, Jin S J, Xu P, Storey K B, Huan P, Zhang T, Zhou Y, Zhang J Q, Lin C G, Li X N, Xing L L, Huo D, Sun M Z, Wang L, Mercier A, Li F H, Yang H S, Xiang J H. 2017. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. Plos Biology, 15(10): e2003790.
DOI:10.1371/journal.pbio.2003790 |
Zhao H, Yang H S, Zhao H L, Chen M Y, Wang T M. 2011. The molecular characterization and expression of heat shock protein 90 (Hsp90) and 26 (Hsp26) cDNAs in sea cucumber (Apostichopus japonicus). Cell Stress and Chaperones, 16(5): 481.
DOI:10.1007/s12192-011-0260-z |
Zhao Y, Chen M Y, Wang T M, Sun L N, Xu D X, Yang H S. 2014. Selection of reference genes for qRT-PCR analysis of gene expression in sea cucumber Apostichopus japonicus during aestivation. Chinese Journal of Oceanology and Limnology, 32(6): 1248-1256.
DOI:10.1007/s00343-015-4004-2 |
 2021, Vol. 39
2021, Vol. 39



