Institute of Oceanology, Chinese Academy of Sciences
Article Information
- HAN Jinfen, NAN Fangru, FENG Jia, LÜ Junping, LIU Qi, LIU Xudong, XIE Shulian
- Affinities of freshwater "Chantransia" stage algae (Rhodophyta) from China based on molecular and morphological analyses
- Journal of Oceanology and Limnology, 39(3): 1063-1076
- http://dx.doi.org/10.1007/s00343-020-0114-6
Article History
- Received Mar. 6, 2020
- accepted in principle Apr. 19, 2020
- accepted for publication Jun. 28, 2020
The freshwater red algal orders Batrachospermales and Thoreales belong to Florideophyceae (Rhodophyta). Freshwater red algal orders Batrachospermales and Thoreales reportedly have triphasic life history, including a macroscopic gametophyte, carposporophytes that develop on the gametophyte thallus, and a "Chantransia" asexual reproduction stage (Sheath, 1984). The morphological characteristics of their "Chantransia" stages are small tufts from a macro perspective and branched filaments under a microscope. However, its simplicity makes this phase a hindrance to species identification. On the one hand, true Audouinella and the "Chantransia" stages of orders Batrachospermales and Thoreales cannot be distinguished due to similar morphological features. Therefore, to solve this problem, researchers proposed that bluish-colored Audouinella-like species manifest the "Chantransia" stage, whereas the reddish-colored species are the true Audouinella species (Skuja, 1934; Necchi et al., 1993a, b). That being said, other studies have confirmed that extrinsic factors cause color changes (Kaczmarczyk and Sheath, 1992). On the other hand, no correlation exists between certain species of orders Batrachospermales and Thoreales and their morphological "Chantransia" stage characteristics. Some researchers have found that "pygmaea" morphology probably represents the "Chantransia" stage of numerous species, and the Batrachospermum antipodites (currently regarded as a synonym of Nocturama antipodites (Entwisle) Entwisle & M.L.Vis in Entwisle et al. 2016: 388, 389) has been associated with two "Chantransia" morphologies (Zucchi and Necchi, 2003; Vis et al., 2006; Necchi and Oliveira, 2011).
Phylogenetic tree reconstructions are traditionally applied as representations of the phylogenetic relationships among species. Currently, the phylogenetic trees were established based on sequences with appropriate evolutionary rates. Studies of molecular sequences of the organellar genes, such as the plastid rbcL, the plastid 23S rRNA gene (UPA), and the mitochondrial COI-5P have made significant contributions to elucidate the relationships within Rhodophyta (e.g., Li et al., 2010; Hind and Saunders, 2013; Nan et al., 2014). The rbcL gene has been analyzed to determine the affinities of fourteen putative "Chantransia" isolates collected from locations in French Guiana, Brazil, and Bolivia from South America, and Mississippi, USA in North America (Chiasson et al., 2005). Phylogenetic analysis showed that "macrospora" isolates were the "Chantransia" of B. macrosporum (currently regarded as a synonym of Montagnia macrospora (Montagne) Necchi, M. L. Vis & A. S. Garcia in Necchi et al. 2019: 587, Figs. 3-14), while the "pygmaea" morphology more likely represents numerous species (Chiasson et al., 2005). In addition, other studies also indicated that A. pygmaea specimens (Kützing, Weber Bosse 1921: 191) were related most closely to B. arcuatum (currently regarded to Sheathia arcuata (Kylin) Salomaki & M.L.Vis in Salomaki et al. 2014: 535) and Nemalionopsis tortuosa (Yoneda & Yagi in Yagi & Yoneda 1940: 85, Figs. 1, 2: 1-8) as based on rbcL sequences (Chiasson et al., 2007). Furthermore, Nan et al. (2016) reported that four Audouinella-like species from China could be classified as Thorea hispida ((Thore) Desvaux 1818: 16) as based on UPA and COI-5P. The purpose of this study was to use morphological and molecular data to determine the phylogenetic positions of the twelve "Chantransia" stage specimens from several regions of China.
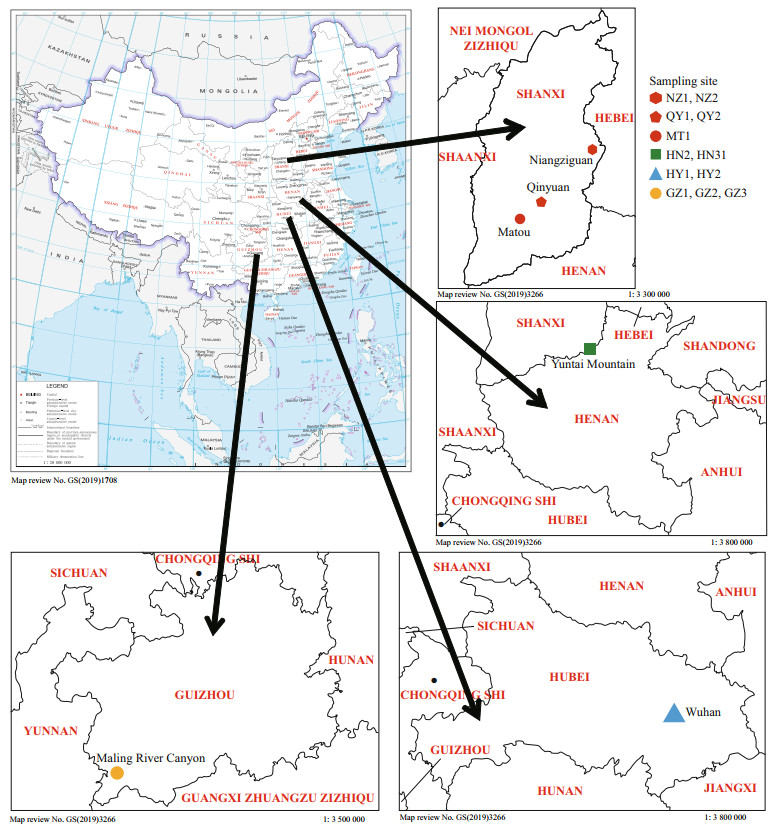
2 MATERIAL AND METHOD 2.1 Sample collectionTwelve freshwater isolates, putatively recognized as "Chantransia", were collected from locations ranging across the Hubei, Shanxi, Guizhou, and Henan Provinces in China. Figure 1 and Table 1 show collection information. Materials were collected using a knife and tweezers and were transferred immediately to the laboratory. Materials were rinsed several times in ultrapure water, removing impurities and epiphytic algae. Specimens for DNA extraction were desiccated in silica gel, whereas the morphological examination material was placed in centrifuge tubes and fixed with a 4% formalin solution.
|
| Fig.1 Map of China (map review No. GS (2019)1708) showing the collection sites of "Chantransia" stage samples investigated in this study More detailed location information is provided in Table 1. |
For morphology observations, both fresh and formalin preserved thalli were examined under a BX- 51 Olympus microscope equipped with a chargecoupled device (DP72; Olympus, Tokyo, Japan) for taking photographs.
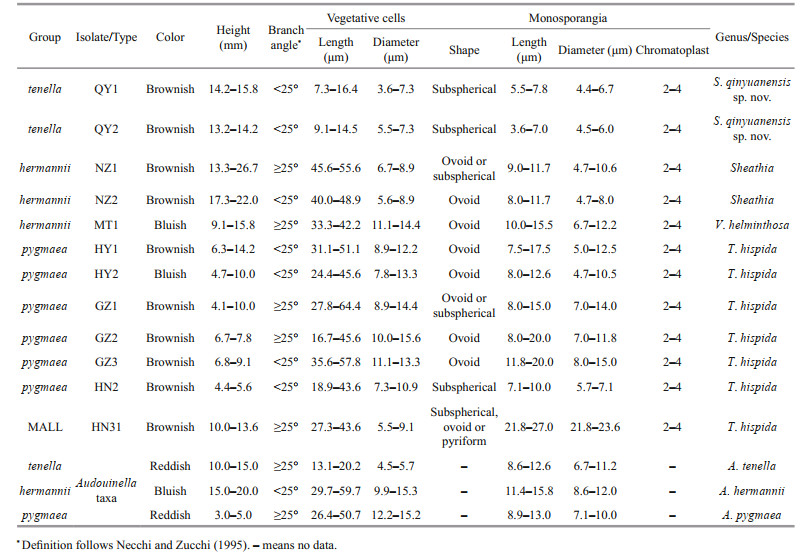
The morphological characteristics of all specimens utilized in this study and three specimens of the genus Audouinella (Table 2) were evaluated using Ward's method of Hierarchical cluster analysis in SPSS 19.0 package (George and Mallery, 2010). The differences among the groups were analyzed by one-way ANOVA via SPSS 19.0 package. P < 0.05 was considered for statistical significance.
|
Total DNA was extracted following the protocol originally described by Saunders (1993) and revised by Vis and Sheath (1997). Polymerase chain reaction amplification reaction volume was 20 μL, consisting of 10.8-μL ddH2O, 2.0-μL 10×buffer, 2.0-μL 2.5-mmol/L dNTPs, 0.2-μL Taq DNA polymerase (all from Sangon Biotech Co., Ltd., China), 2.0 μL of each primer (10 mmol/L), and 1.0 μL of genomic DNA. The reaction was executed in a MyCycler thermal cycler (Bio-Rad, USA). rbcL gene was amplified using the primer pair F160 (5′-CCTCAACCAGGAGTAGAT-3′) and rbcLR (5′-ACATTTGCTGTTGGAGTCTC-3′) (Vis et al., 1998). Also, another pair of primers, F650 (5′-ATTAACTCTCAACCATTTATGCG-3′) and rbcLR (Salomaki et al., 2014) were the substitutive primers so long as the first pair did not produce a PCR product. For rbcL, the PCR procedure consisted of an initial denaturation at 95 ℃ for 2 min, 35 cycles of denaturation at 93 ℃ for 1 min, annealing at 47 ℃ for 1 min, extension at 72 ℃ for 2 min, and a final extension of 2 min at 72 ℃. The primer p23SrV_f1 (5′-GGACAGAAAGACCCTATGAA-3′) and p23SrV_r1 (5′-TCAGCCTGTTATCCCTAGAG-3′) (Sherwood and Presting, 2007) were utilized to amplify UPA sequences. PCR cycling conditions for UPA were as follows: 2 min initial denaturation at 94 ℃, 35 cycles at 94 ℃ for 20 s, 55 ℃ for 30 s, 72 ℃ for 30 s, and a final extension at 72 ℃ for 10 min. The primers GazF1 (5′-TCAACAAATCATAAAGATATTGG-3′) and GazR1 (5′-ACTTCTGGATGTCCAAAAAAYCA-3′) (Saunders, 2005) were utilized to amplify COI-5P. For COI-5P, thermal cycling conditions included an initial denaturation step at 94 ℃ for 4 min, 35 cycles at 94 ℃ for 1 min, 45 ℃ for 30 s, 72 ℃ for 1 min, and a final extension at 72 ℃ for 7 min.
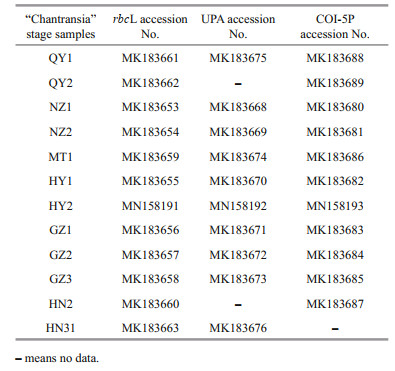
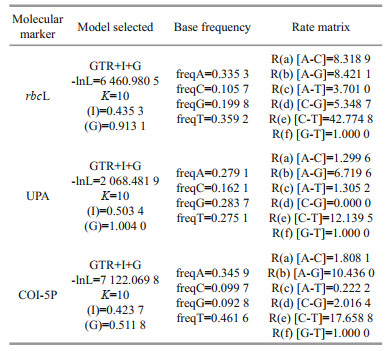
2.4 Sequence determination and data analysisPCR products, along with their amplification primers, were sent to the BGI Tech Corporation (Beijing, China) for sequencing on an ABI 3730XL sequencer. Sequences generated in this study were deposited in GenBank (Table 3). The other Batrachospermales, Thoreales, and Audouinella taxa sequences utilized in this study for analysis were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) (listed in Supplementary Table S1), and two genera, Compsopogon and Bangia, were incorporated as outgroup taxa. DNA sequence data produced by this study were aligned with sequences obtained from GenBank using Clustal-X 2.0 (Thompson et al., 1997) and MEGA 5.0 (Tamura et al., 2011). Untrimmed bases on both ends were deleted to produce identical length alignment. Furthermore, MEGA 5.0 was utilized to calculate the pairwise sequence distances. Phylogenetic trees were constructed based on the aligned sequences. Appropriate models of sequence evolution and related parameters of the rbcL, UPA, and COI-5P gene sequences datasets were generated by adopting Modeltest 3.7 (Posada and Buckley, 2004) with the results listed in Table 4. MEGA 5.0 was utilized to establish Neighbor-joining (NJ) trees with 1 000 bootstrap replications. PHYML software (Felsenstein, 1981; Guindon and Gascuel, 2003) was utilized to construct the Maximum Likelihood (ML) trees with 1 000 replicates of bootstrap analysis. Meanwhile, Bayesian Inferences (BI) was developed in MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003). The Markov chain Monte Carlo (MCMC) algorithm was utilized to establish 4 Markov chains. Three hot chains, 1 cold chain, and 5 million generations sampling every 1 000 generations were employed. The burn-in for each run, determined by plotting the overall likelihood scores against the generation, was set as 25 ℃ of the total tree by default. Resulting phylogenetic trees were edited using Figtree1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
|
|
Morphometric data showed a wide range in all characteristics among isolates. Hierarchical cluster analysis grouped these specimens and three species of genus Audouinella into four clusters (Fig. 2).

|
| Fig.2 The cluster relationship of 12 putative "Chantransia" isolates and three specimens of freshwater Audouinella showing four groupings |
The first group (named tenella) included two isolates (QY1 and QY2) in this study, as well as a species of genus Audouinella (A. tenella (Skuja) Papenfuss 1945: 326). Specimens of this group are distinguished as macroscopic (13.2-15.8 mm), having small vegetative cells ((7.3-16.4) μm×(3.6-7.3) μm), and monosporangia ((4.4-6.7) μm in diameter) (Table 2; Fig. 3a & b).

|
| Fig.3 Morphological features of putative "Chantransia" isolates in this study a. isolate QY1 (=Sheathia qinyuanensis sp. nov.). Monosporangial branch with subspherical monosporangium (arrowhead); b. isolate QY2 (=Sheathia qinyuanensis sp. nov.). Monosporangial branch with subspherical monosporangium (arrowhead); c. isolate NZ1. Filament showing branch angle ≥25° (arrow) and monosporangium (arrowhead); d. isolate NZ2. Filament showing branch angle ≤25° (arrow) and monosporangium (arrowhead); e. isolate MT1. Filament showing branch angle ≥25° (arrow) and monosporangia (arrowhead); f. isolate HY1. Monosporangium (arrowhead) grow binately on top of the spore branch; g. isolate HY2. Filament with a monosporangial branch and an ovoid monosporangium (arrowhead); h. isolate GZ1. Monosporangial branch with monosporangium (arrowhead); i. isolate GZ2. Filament showing branch angle ≥25° (arrow) and monosporangial branch with a mature monosporangium (arrowhead) and two empty, discharged monosporangium (double arrowheads); j. isolate GZ3. Filament showing branch angle ≤25° (arrow) and monosporangia (arrowhead); k. isolate HN2. Monosporangial branch with subspherical monosporangium (arrowhead); l. isolate HN31. Monosporangial branch with big subspherical (arrow), ovoid (arrowhead) or pyriform monosporangia (double arrowheads). Scale bar: 20 μm (a-l). |
The second group (named hermannii) consisted of three isolates (NZ1, NZ2, and MT1), as well as a species of genus Audouinella (A. hermannii (Roth) Duby 1830: 972). This group is characterized as macroscopic (9.1-26.7 mm), having large vegetative cells ((29.7-59.7) μm×(5.6-15.3) μm), and small monosporangia ((4.7-12.2) μm in diameter) (Table 2; Fig. 3c-e).
The third large group (called pygmaea) contained six isolates (HY1, HY2, GZ1, GZ2, GZ3, and HN2), as well as a species of genus Audouinella (A. pygmaea). It is separated from the other groups as microscopic (4.1-14.2 mm), having large vegetative cells ((16.7- 64.4) μm×(7.3-15.6) μm), and small monosporangia ((4.7-15.0) μm in diameter) (Table 2; Fig. 3f-k).
The fourth group (called MALL) included one isolate (HN31) and no Audouinella taxa. It is distinct from other groups as macroscopic (10.0-13.6 mm), having large vegetative cells ((27.3-43.6) μm×(5.5- 9.1) μm), and monosporangia ((21.8-23.6) μm in diameter) (Table 2; Fig. 3l).
Isolate HN2 had the smallest mean tuft length (5.0 mm) and isolate NZ1 had the largest (20.0 mm), with their overall range being 13.3-26.7 mm. The mean tuft length significantly differed (P < 05) from each group, and only the isolates belonging to groups "tenella" and "MALL" were similar. Isolates of group "hermannii" were significantly larger (P < 05) than others regarding mean tuft length and vegetative cell length, but they did not differ from at least some of the other isolates in vegetative cell diameter and monosporangia size. The mean vegetative cell diameter significantly differed (P < 05) among the isolates belonging to groups "pygmaea", "tenella", and "MALL". Isolates belonging to groups "hermannii" and "pygmaea" were quite consistent regarding monosporangia size, whereas isolates of group "tenella" were significantly smaller (P < 05) than all other isolates regarding monosporangia length, and isolate HN31 was significantly larger (P < 05) than the other isolates regarding monosporangia length and diameter (Table 2).
3.2 Molecular analysis 3.2.1 Analysis of rbcL sequencesrbcL data matrix included 46 specimens of two orders Batrachospermales and Thoreales and 3 species of a phylogenetically distant genus Audouinella, as well as 2 outgroup taxa, consisting of 644 characters, of which 295 (45.81%) were variable and 272 (42.24%) were parsimony informative. Supplementary Table S2 lists the pairwise distance among "Chantransia" isolates and other specimens of orders Batrachospermales and Thoreales.
Table 2 lists the pairwise distance based on rbcL sequences between all 12 "Chantransia" isolates and other species of orders Batrachospermales and Thoreales. Results show that isolate MT1 was identical to Virescentia helminthosa (Bory) Necchi, D. C. Agostinho & M. L. Vis 2018: 318. However, the p-distance between isolates QY1 and QY2 was 1%, corresponding to 8 single-bp differences. The pairwise distance between isolates collected from Qinyuan of Shanxi Province (QY1 and QY2) and the other species of genus Sheathia showed larger variance than among intraspecific distance of the genus Sheathia (0.0582 vs 0.0565). The p-distances among isolates NZ1, NZ2, and four species of genus Sheathia (S. hongdongensis (S. L. Xie & J. Feng) J. -F. Han et al. 2018: 68, S. jinchengensis J. -F. Han et al. 2018: 65, Fig. 1, S. longipedicellata (Hua & Shi) J. -F. Han et al. 2018: 68, and S. matouensis J. -F. Han et al. 2019: 258, Fig. 2) were 0%-1%. Similarly, divergences between the remaining seven isolates used in this study and T. hispida specimens (including both the gametophyte and "Chantransia" stage) were 0%-1%.
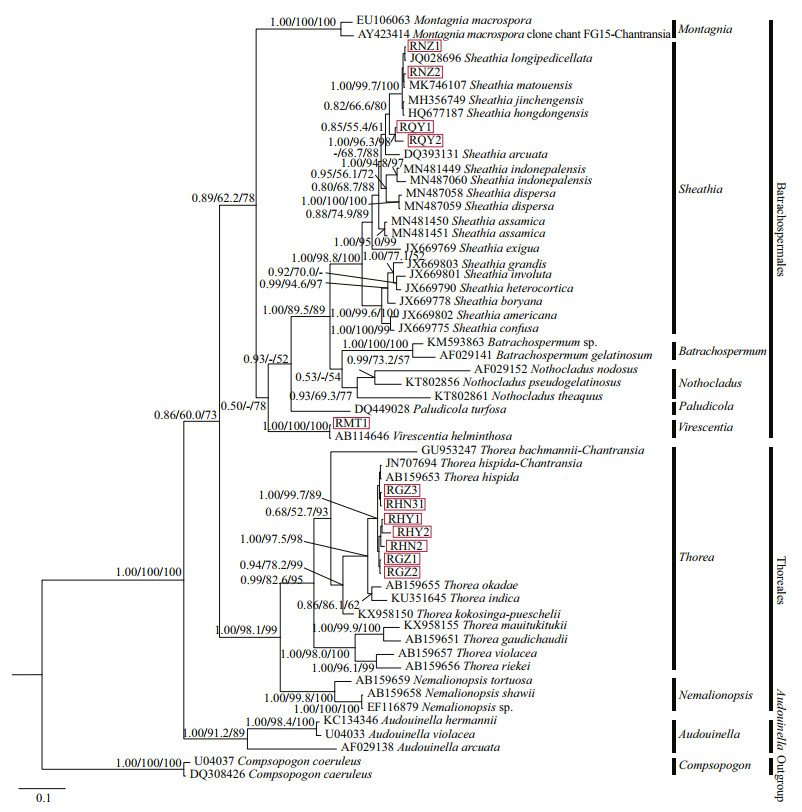
Three phylogenetic methods (NJ, BI, and ML) produced trees with similar topological structures among "Chantransia" isolates and their sister specimens, except that the main branches were arranged differently. Therefore, only the BI tree with supporting values calculated from three methods was displayed (Fig. 4).
|
| Fig.4 Bayesian inference tree based on the rbcL gene sequences Support values for all analyses are shown as follows: Bayesian posterior probabilities/ML bootstrap/NJ distance bootstrap. '-' denotes < 50% support for that analyses at that node. All new sequences generated in this study are indicated in red boxes. |
rbcL tree was composed of three main branches representing orders Batrachospermales, Thoreales, and genus Audouinella. In the Batrachospermales branch, four putative "Chantransia" isolates clustered with Sheathia taxa that had high support values (1.00/98.8/100). Isolates collected from Qinyuan of Shanxi Province (QY1 and QY2) clustered into an independent clade. However, they were separated from other species of genus Sheathia with moderate Bayesian posterior probabilities (PP) and NJ support values (0.82/66.6/80). Among the Sheathia taxa, four species S. hongdongensis, S. jinchengensis, S. longipedicellata, and S. matouensis showed the closest relationship with isolates NZ1 and NZ2, which was well-supported (1.00/99.7/100). Isolate MT1 was also in the Batrachospermales branch and clustered with V. helminthosa that had robust values (1.00/100/100). Within the Thorea branch, the seven other "Chantransia" isolates and T. hispida clustered together with moderate to high support values (1.00/99.7/89). They were sister to a clade formed by a species of T. okadae (Yamada 1949: 158, Figs. 1-3 (as 'okadai')) and T. indica (Necchi, E. K. Ganesan & J. A. West 2015: 268, Fig. 2A-I). However, none of the 12 isolates clustered with Audouinella algae.
3.2.2 Analysis of UPA sequencesThe UPA sequence determined for the 10 (except for isolates HN2 and QY2) "Chantransia" isolates utilized in this study were 310 bp, of which 115 (37.10%) were variable and 99 (31.94%) were parsimony informative. Supplementary Table S3 lists the pairwise distance among "Chantransia" isolates and other specimens of orders Batrachospermales and Thoreales. For the UPA gene, three different phylogenetic methods produced trees in similar topology. Therefore, only the BI tree was shown indicating supporting values calculated from the three methods in Fig. 5.

|
| Fig.5 Bayesian inference tree based on the UPA gene sequences Support values for all analyses are shown as follows: Bayesian posterior probabilities/ML bootstrap/NJ distance bootstrap. '-' denotes < 50% support for that analyses at that node. All new sequences generated in this study are indicated in red boxes. |
Isolate MT1 was located in the Batrachospermales branch and formed a moderately-to-strongly supported clade with the Virescentia species (1.00/83.2/85). Divergence within this clade was 4%, corresponding to 11-bp differences. Furthermore, within the Sheathia clade, three putative "Chantransia" isolates formed two independent clades that roughly related to the gathering places: the first clade was composed of isolates NZ1 and NZ2 collected from Niangziguan of Shanxi Province. They clustered together with moderate-to-high support values (0.95/81.0/85) and were sister to a clade formed by species of S. hongdongensis and S. boryana ((Sirodot) Salomaki & M. L. Vis in Salomaki et al. 2014: 535), while isolate QY1, collected from Qinyuan of Shanxi Province, formed the second weakly supported independent clade. The remaining six samples (GZ1, GZ2, GZ3, HY1, HY2, and HN31) in this study were in the Thoreales clade. Among the Thorea species, T. hispida demonstrated the closest relationship with these six "Chantransia" isolates, which was well supported by the p-distance (0%-2%). However, in the UPA tree, these six isolates clustered together with T. hispida that only had high support values for NJ (-/66.9/99).
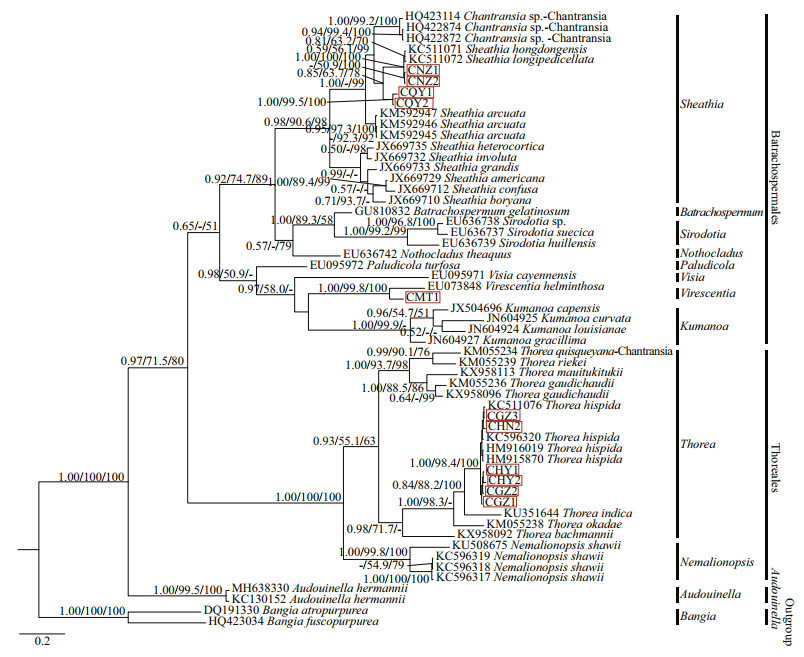
3.2.3 Analysis of COI-5P sequencesIn this study, the COI-5P alignment of species was 573 bp, with 287-bp variable sites (among which 265 bp were Parsimony-Informative sites). Supplementary Table S4 lists the pairwise distance between the "Chantransia" isolates and other specimens of orders Batrachospermales and Thoreales. All three phylogenetic analyses (NJ, BI, and ML) based on COI-5P alignments showed similar topological structures. Therefore, only the BI trees were shown indicating supporting values calculated from the three methods in Fig. 6.
|
| Fig.6 Bayesian inference tree based on the COI-5P gene sequences Support values for all analyses are shown as follows: Bayesian posterior probabilities/ML bootstrap/NJ distance bootstrap. '-' denotes < 50% support for that analyses at that node. All new sequences generated in this study are indicated in red boxes. |
Pairwise distance based on COI-5P sequences showed that sequence divergences among MT1 and other species of the order Batrachospermales were 16%-21%, which were much larger than the variation between MT1 and V. herminthosa (9%). Meanwhile, two identical isolates, NZ1 and NZ2, differed from S. hongdongensis and S. longipedicellata at a single nucleotide position. Similar to results based on the rbcL sequences, isolates (QY1 and QY2) collected from Qinyuan of Shanxi Province had unique sequences, and the distances between them and the other species of genus Sheathia were larger than the intraspecific distances of this genus (0.112 0 vs 0.092 8). Furthermore, divergences between the remaining six isolates utilized in this study and the specimens of T. hispida were only 0%-2%, corresponding to 0-11-bp differences, while the average distance between these 6 isolates in the Thoreales branch and the other species of genus Thorea was 14%.
All in-group species were clustered into three main branches, corresponding to Batrachospermales, Thoreales, and Audouinella. Within the Batrachospermales branch, isolate MT1 clustered with V. helminthosa and formed a strongly supported clade (1.00/99.8/100). The four other putative "Chantransia" isolates NZ1, NZ2, QY1, and QY2 clustered together with species of genus Sheathia and formed a monophyletic clade that had high support values (0.98/90.6/98). Meanwhile, isolates NZ1 and NZ2 clustered with S.hongdongensis and S.longipedicellata that had high support values (1.00/100/100), while the other two isolates (QY1 and QY2) collected from Qinyuan of Shanxi Province formed an independent clade. However, they were separated from other species of genus Sheathia with moderate support values for PP and NJ (0.85/63.7/78). Moreover, the remaining six samples (GZ1, GZ2, GZ3, HY1, HY2, and HN2) in this study, together with previously reported species of genus Thorea, clustered with genus Nemalionopsis to form the Thoreales branch. In this branch, isolates GZ1, GZ2, GZ3, HY1, HY2, and HN2 clustered in a single clade together with T. hispida that had high support values (1.00/98.4/100).
4 DISCUSSIONDespite the research on the delimitation of Acrochaetiaceae having been executed for a long time, some controversy remains one aspect of which is the applicability of criteria proposed by Skuja (1934). Based on his proposal, all reddish species morphologically similar to Audouinella are true Audouinella, while bluish species represent the "Chantransia" stage. However, several subsequent studies have queried this conclusion. For example, Kaczmarczyk and Sheath in 1992 clarified that freshwater red algae color change is related to irradiance (Kaczmarczyk and Sheath, 1992). Additionally, Chen et al. (2014) reported color changes (bluish-green in the stream that turned reddish in light) of an acrochaetioid algae affected by light. These results, supported by both morphological and molecular data, show that all 12 isolates that are reddish, bluish, and brownish belong to the "Chantransia" stages of orders Batrachospermales and Thoreales. Consequently, when combined with previous studies, caution is recommended in thallus color criteria which were considered a standard for identifying true Audouinella, combining molecular analysis with morphological methods is essential.
Owing to the limitations of morphological methods when analyzing the affinities of putative "Chantransia" stages, molecular-assisted methods have been used extensively (Harper and Saunders, 1998; Pueschel et al., 2001). Similar to A. macrospora ((Wood) Sheath & Burkholder 1985: 111 (Table 1)), isolate HN31 has large monosporangia (>20 μm in diameter) (Table 2) (Necchi and Zucchi, 1997). In previous studies, all "A. macrospora" is indicated to be the "Chantransia" of B. macrosporum (Pueschel et al., 2001; Chiasson et al., 2005). However, molecular phylogeny obtained in this study unequivocally demonstrated that isolate HN31 was the "Chantransia" stage of T. hispida. In addition to isolate HN31, this study also confirmed that six "pygmaea" isolates (GZ1, GZ2, GZ3, HY1, HY2, and HN2) collected from the Guizhou, Hubei, and Henan Provinces also belong to T. hispida. Considering the locations of T. hispida that were previously reported in China, including Shanxi, Jiangsu, Yunnan, and Guizhou Provinces, the number of T. hispida distribution sites is proposed to have expanded to six (Shi, 2006). Pairwise distance and molecular phylogenetic trees both supported that one of the three "hermannii" isolates MT1 can be assigned to V. helminthosa, while the other two "hermannii" isolates NZ1 and NZ2 formed a lineage and were allied with several Sheathia taxa. Furthermore, a new species of genus Sheathia, corresponding to two "A. tenella" morphologies (QY1, QY2), is proposed primarily based on the DNA sequence data generated in this study, otherwise known as S. qinyuanensis.
Sheathia qinyuanensis HAN, NAN ET XIE sp. nov. (Fig. 3a & b)
Known only from the "Chantransia" sporophyte generation. Tuft-shape, with the thallus being brownish, 13.2-15.8 mm length, with erect and densely branched filaments, and branch angle < 25°. Vegetative cells are cylindrical, 7.3-16.4 μm in length and 3.6-7.3 μm in diameter, reproducing via abundant ovoid or sub-spherical monosporangia, 3.6-7.8 μm in length and 4.4-6.7 μm in diameter.
Etymology—The species epithet refers to the type locality (Qinyuan, China).
Type—China, Shanxi Province, Qinyuan (36°29′N, 112°19′E): on the surface of the stone in flowing water, May 2018, Fang-Ru NAN (Holotype: SXUQY18051; Paratype: SXU-QY18052). It is deposited in the Herbarium of Shanxi University (SXU), Shanxi University, Taiyuan, Shanxi Province, China.
Authentic strain—SXU-QY18051.
Representative DNA Barcode: GenBank MK183661 and MK183662 for rbcL, MK183675 for UPA, and MK183688 and MK183689 for COI.
The red algal genus Thorea is classified in the Rhodophyta (Florideophyceae, Thoreales, Thoreaceae). Until now, sixteen species have been reported worldwide. However, only two species (T. hispida and T. violacea Bory 1808: 133, pl. 18: Fig. 2) have been reported in China (Xie and Shi, 2003). Also, some research has confirmed that T. hispida is associated with several different morphological "Chantransia" stages. Based on seven gene markers, Han (2012) performed a phylogenetic analysis of A. sinensis (C.-C.Jao 1940: 241, pl. 1: Figs. 5 & 6) and A. heterospora (S. L. Xie & Y. J. Ling 1998: 281, Fig. 1) and found that they are actually "Chantransia" of T. hispida. On the bases of rbcL and SSU rDNA, Chen et al. (2014) found that samples morphologically identified as A. heterospora represent the "Chantransia" stage of T. hispida. Furthermore, Nan et al. (2016) reported that four Audouinella-like species (both reddish and bluish thalli included) were the "Chantransia" stage of T. hispida. Combined with the results of this study, T. hispida is concluded to be associated with at least four "Chantransia" morphologies ("A. sinensis", "A. heterospora", "A. pygmaea", and the specimens morphologically similar to the isolate HN31 in this study).
Order Batrachospermales belongs to class Florideophyceae (Rhodophyta), which included most freshwater red algae (Entwisle et al., 2009). In this order, the genus Batrachospermum was divided into several sections according to morphological characteristics (Kumano, 2002). However, molecular phylogenetic analysis has confirmed that some sections of this genus are monophyletic and can be separated from genus Batrachospermum (Entwisle et al., 2009; Vis et al., 2012; Salomaki et al., 2014; Necchi et al., 2019). Genus Virescentia was elevated from one section (section Virescentia Sirodot) of the genus Batrachospermum (Necchi et al., 2018). Until now, six species of this genus have been reported worldwide, of which V. helminthosa is the holotype species (Guiry and Guiry, 2020). In China, up to this point in time, this new genus has not been extensively studied, while section Virescentia has been studied in a few reports (Xie, 2001; Ji, 2013). According to morphometric identification, B. helminthosum (currently regarded as a synonym of V. helminthosa) was reported from Zhejiang and Chongqing (Jao, 1941). In this study, isolate MT1, which is morphologically similar to A. hermannii, was collected from Matou of Shanxi Province and was concluded to represent the "Chantransia" stage of V. helminthosa. Consequently, the number of V. helminthosa distribution sites in China expands to three.
Traditional classification methods are mainly based on the morphological characteristics of algae. However, the morphological characteristics of "Chantransia" stages will change under the influence of factors such as physiology and ecology. Nucleotides are carriers of genetic information; therefore evaluating the evolutionary relationship of species via the homology of nucleotide sequences is extremely important. Ji et al. (2017) assessed DNA barcode markers for freshwater red algae and suggested that four sequence regions (COI-5P, cox2-3 spacer, UPA, and rbcL) were useful for species-level identification in genus Batrachospermum, except for some allied species. rbcL marker was utilized by Chiasson et al. (2005) and by Necchi and Oliveira (2011) for identifying "Chantransia" isolates in members of Batrachospermales and Thoreales, strongly confirming that all "macrospora" isolates were "Chantransia" of B. macrosporum, whereas "C. pygmaea" was associated with at least nine species. Also, molecular sequences of COI-5P and UPA were utilized to assess phylogenetic positions of four Audouinella-like species from China, showing high species identification resolution (Nan et al., 2016). Johnston et al. (2018) reported a new species—Thorea quisqueyana (E. T. Johnston & M. L. Vis)—based on its "Chantransia" stage samples and three DNA sequences (rbcL, COI- 5P, and LSU). Based on the phylogenetic analysis of rbcL and the COI-5P gene, a new genus Virescentia and a new species V. viride-americana (Necchi, D. C. Agostinho & M. L. Vis 2018: 323, Figs. 18-22) were proposed by Necchi et al. (2018). Moreover, another new genus Montagnia was also proposed that was based on rbcL and COI-5P sequence data (Necchi et al., 2019). Furthermore, much research revealed that the UPA gene can distinguish samples at the species level, even though the sequence is shorter and the intra- and inter-specific divergence values are relatively lower (Sherwood and Presting, 2007; Clarkston and Saunders, 2010). In this study, two chloroplast genes (rbcL and UPA) and a mitochondrial gene (COI-5P) were used to elucidate the relationships between all twelve "Chantransia" isolates and the species of Audouinella, Batrachospermales, and Thoreales. Twelve isolates were demonstrated to belong unequivocally to the "Chantransia" stages of orders Batrachospermales and Thoreales. Moreover, many species of freshwater red algae, especially orders Batrachospermales and Thoreales, may exist in the "Chantransia" stage for a long time when environmental conditions are unsuitable for gametophyte growth. When compared with two longer molecular markers, SSU and LSU, utilized by Chen et al. (2014) and Johnston et al. (2018), the three DNA markers used in this study (rbcL, UPA, and COI-5P) are more rapid and convenient for species identification.
5 CONCLUSIONBased on all three DNA fragments, including rbcL, UPA, and COI-5P, all twelve isolates in this study were demonstrated to belong unequivocally to three different genera Sheathia, Virescentia, and Thorea, corresponding to the two orders Batrachospermales and Thoreales. The proposal of S. qinyuanensis as a new species of genus Sheathia from China is supported by DNA sequence data generated in this study. Furthermore, isolate MT1 was demonstrated to be the "Chantransia" of V. helminthosa, which expands the V. helminthosa species distribution in China to three. Seven isolates (GZ1, GZ2, GZ3, HY1, HY2, HN2, and HN31) represented the "Chantransia" of T. hispida, of which the descriptions of HY1, HY2, HN2, and HN31 expand the T. hispida species distribution in China to six.
6 DATA AVAILABILITY STATEMENTThe data that support the findings of this study are available on request from the corresponding author (email: xiesl@sxu.edu.cn).
7 ACKNOWLEDGMENTDr. S. James (University of California, Davis) is acknowledged for the English editing. We sincerely thank Prof. LIU Guoxiang from Institute of Hydrobiology, Chinese Academy of Sciences for providing samples. Dr. ZHU Huan from Institute of Hydrobiology, Chinese Academy of Sciences is acknowledged for providing information on the sample collecting.
Electronic supplementary materialSupplementary material (Supplementary Tables S1–S4) is available in the online version of this article at https://doi.org/10.1007/s00343-020-0114-6.
Chen L, Feng J, Han X J, Xie S L. 2014. Investigation of a freshwater acrochaetioid alga (Rhodophyta) with molecular and morphological methods. Nordic Journal of Botany, 32(5): 529-535.
DOI:10.1111/njb.00407 |
Chiasson W B, Johanson K G, Sherwood A R, Vis M L. 2007. Phylogenetic affinities of form taxon Chantransia pygmaea (Rhodophyta) specimens from the Hawaiian Islands. Phycologia, 46(3): 257-262.
DOI:10.2216/06-79.1 |
Chiasson W B, Sabo N J, Vis M L. 2005. Affinities of freshwater putative chantransia stages (Rhodophyta) from molecular and morphological data. Phycologia, 44(2): 163-168.
DOI:10.2216/0031-8884(2005)44[163:AOFPCS]2.0.CO;2 |
Clarkston B E, Saunders G W. 2010. A comparison of two DNA barcode markers for species discrimination in the red algal family Kallymeniaceae (Gigartinales, Florideophyceae), with a description of Euthora timburtonii sp. nov. Botany, 88(2): 119-131.
DOI:10.1139/B09-101 |
Entwisle T J, Vis M L, Chiasson W B, Necchi O Jr, Sherwood A R. 2009. Systematics of the Batrachospermales (Rhodophyta)-a synthesis. Journal of Phycology, 45(3): 704-715.
DOI:10.1111/j.1529-8817.2009.00686.x |
Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution, 17(6): 368-376.
DOI:10.1007/BF01734359 |
George D, Mallery P. 2010. SPSS for Windows Step By Step: A Simple Guide and Reference, 17.0 Update. 10th edn. Allyn & Bacon, Boston. 386p.
|
Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52(5): 696-704.
DOI:10.1080/10635150390235520 |
Guiry M D, Guiry G M. 2020. World-wide electronic publication, National University of Ireland, Galway. AlgaeBase, http://www.algaebase.org. Accessed on 2020-01-13.
|
Han X J. 2012. Phylogenetic Relationship of Audouinella Based on Gene Sequences. Master's dissertation, Shanxi University. 65p. (in Chinese with English abstract)
|
Harper J, Saunders G. 1998. A molecular systematic investigation of the Acrochaetiales (Florideophycidae, Rhodophyta) and related taxa based on nuclear small-subunit ribosomal DNA sequence data. European Journal of Phycology, 33(3): 221-229.
DOI:10.1080/09670269810001736723 |
Hind K R, Saunders G W. 2013. A molecular phylogenetic study of the Tribe Corallineae (Corallinales, Rhodophyta) with an assessment of genus-level taxonomic features and descriptions of novel genera. Journal of Phycology, 49(1): 103-114.
DOI:10.1111/jpy.12019 |
Jao C C. 1941. Studies on the freshwater algae of China. Ⅷ. A preliminary account of the Chinese freshwater rhodophyceae. Sinensia, 12: 245-290.
|
Ji L, Feng J, Nan F R, Chen L, Hu B F, Xie S L. 2017. Evaluating DNA barcode markers for freshwater red algae: a case study using family Batrachospermaceae. Acta Hydrobiologica Sinica, 41(3): 643-651.
(in Chinese with English abstract) DOI:10.7541/2017.82 |
Ji L. 2013. Molecular Phylogenetics of Batrachospermum Roth (Rhodophyta) of China. Doctoral dissertation, Shanxi University. 197p. (in Chinese with English abstract)
|
Johnston E T, Dixon K R, West J A, Buhari N, Vis M L. 2018. Three gene phylogeny of the Thoreales (Rhodophyta) reveals high species diversity. Journal of Phycology, 54(2): 159-170.
DOI:10.1111/jpy.12618 |
Kaczmarczyk D, Sheath R G. 1992. Pigment content and carbon to nitrogen ratios of freshwater red algae growing at different light levels. Japanese Journal of Phycology, 40: 279-282.
|
Kumano S. 2002. Freshwater Red Algae of the World. Biopress Ltd, Bristol. 375p.
|
Li Q, Ji L, Xie S L. 2010. Phylogenetic analysis of Batrachospemales (Florideophyceae, Rhodophyta) based on chloroplast rbcL sequences. Acta Hydrobiologica Sinica, 36(1): 20-28.
DOI:10.3724/SP.J.1035.2010.00020 |
Nan F R, Feng J, Han X J, Lv J P, Liu Q, Xie S L. 2016. Molecular identification of Audouinella-like species (Rhodophyta) from China based on three short DNA fragments. Phytotaxa, 246(2): 107-119.
DOI:10.11646/phytotaxa.246.2.2 |
Nan F R, Feng J, Xie S L. 2014. Phylogenetic relationship of genus Kumanoa (Batrachospermales, Rhodophyta) based on chloroplast UPA genes. Bulletin of Botanical Research, 34(1): 584-591.
DOI:10.7525/jissn.1673-5102.2014.05.002 |
Necchi O Jr, Agostinho D C, Vis M L. 2018. Revision of Batrachospermum section Virescentia (Batrachospermales, Rhodophyta) with the establishment of the new genus, Virescentia stat. nov.. Cryptogamie Algologie, 39(3): 313-338.
DOI:10.7872/crya/v39.iss3.2018.313 |
Necchi O Jr, Filho A G, Paiano M O. 2019. Revision of Batrachospermum sections Acarposporophytum and Aristata (Batrachospermales, Rhodophyta) with the establishment of the new genera Acarposporophycos and Visia. Phytotaxa, 395(2): 51-65.
DOI:10.11646/phytotaxa.395.2.1 |
Necchi O Jr, Fo A S G, Paiano M O, Vis M L. 2019. Revision of Batrachospermum section Macrospora (Batrachospermales, Rhodophyta) with the establishment of the new genus Montagnia. Phycologia, 58(6): 582-591.
DOI:10.1080/00318884.2019.1624143 |
Necchi O Jr, Oliveira M C. 2011. Phylogenetic affinities of "Chantransia" stages in members of the Batrachospermales and Thoreales (Rhodophyta). Journal of Phycology, 47(3): 680-686.
DOI:10.1111/j.1529-8817.2011.00997.x |
Necchi O Jr, Sheath R G, Cole K M. 1993a. Systematics of freshwater Audouinella (Acrochaetiacees, Rhodophyta) in North America. 1. The reddish species. Algological Studies, 70: 11-28.
|
Necchi O Jr, Sheath R G, Cole K M. 1993b. Systematics of freshwater Audouinella (Acrochaetiacees, Rhodophyta) in North America. 2. The bluish species. Algological Studies, 71: 13-21.
|
Necchi O Jr, Zucchi M R. 1995. Systematics and distribution of freshwater Audouinella (Acrochaetiaceae, Rhodophyta) in Brazil. European Journal of Phycology, 30(3): 209-218.
DOI:10.1080/09670269500650991 |
Necchi O Jr, Zucchi M R. 1997. Audouinella macrospora (Acrochaetiaceae, Rhodophyta) is the chantransia stage of Batrachospermum (Batrachospermaceae). Phycologia, 36(3): 220-224.
DOI:10.2216/i0031-8884-36-3-220.1 |
Posada D, Buckley T R. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology, 53(5): 793-808.
DOI:10.1080/10635150490522304 |
Pueschel C M, Saunders G W, West J A. 2001. Affinities of the freshwater red alga Audouinella macrospora (Florideophyceae, Rhodophyta) and related forms based on SSU rRNA gene sequence analysis and pit plug ultrastructure. Journal of Phycology, 36(2): 433-440.
DOI:10.1046/j.1529-8817.2000.99173.x |
Ronquist F, Huelsenbeck J P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1 572-1 574.
DOI:10.1093/bioinformatics/btg180 |
Salomaki E D, Kwandrans J, Eloranta P, Vis M L. 2014. Molecular and morphological evidence for Sheathia gen. nov. (Batrachospermales, Rhodophyta) and three new species. Journal of Phycology, 50(3): 526-542.
DOI:10.1111/jpy.12179 |
Saunders G W. 1993. Gel purification of red algal genomic DNA: an inexpensive and rapid method for the isolation of polymerase chain reaction-friendly DNA. Journal of Phycology, 29(2): 251-254.
DOI:10.1111/j.0022-3646.1993.00251.x |
Saunders G W. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philosophical Transactions of the Royal Society B: Biological Sciences, 360(1462): 1 879-1 888.
DOI:10.1098/rstb.2005.1719 |
Sheath R G. 1984. The biology of freshwater red algae. In: Round F E, Chapman D J eds. Progress in Phycological Research, vol. 3. Biopress Ltd, Bristol, UK. p. 89-157.
|
Sherwood A R, Presting G G. 2007. Universal primers amplify a 23S rDNA plastid marker in eukaryotic algae and cyanobacteria. Journal of Phycology, 43(3): 605-608.
DOI:10.1111/j.1529-8817.2007.00341.x |
Shi Z X. 2006. Flora Algarum Sinicarum Aquae Dulci, Tomus XⅢ, Rhodophyta, Phaeophyta. Science Press, Beijing, China. p. 1-77.
(in Chinese)
|
Skuja H. 1934. Untersuchungen über die Rhodophyceen des Süsswassers. Botanisches Centralblatt, Beiheft, 52: 173-192.
|
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10): 2 731-2 739.
DOI:10.1093/molbev/msr121 |
Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. 1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25(24): 4 876-4 882.
DOI:10.1093/nar/25.24.4876 |
Vis M L, Entwisle T J, West J A, Ott F D. 2006. Ptilothamnion richardsii (Rhodophyta) is a Chantransia stage of Batrachospermum. European Journal of Phycology, 41(1): 125-130.
DOI:10.1080/09670260500418504 |
Vis M L, Necchi O Jr, Chiasson W B, Entwisle T J. 2012. Molecular phylogeny of the genus Kumanoa (Batrachospermales, Rhodophyta). Journal of Phycology, 48(3): 750-758.
DOI:10.1111/j.1529-8817.2012.01141.x |
Vis M L, Saunders G W, Sheath R G, Dunse K, Entwisle T J. 1998. Phylogeny of the Batrachospermales (Rhodophyta) inferred from rbcL and 18S ribosomal DNA gene sequences. Journal of Phycology, 34(2): 341-350.
DOI:10.1046/j.1529-8817.1998.340341.x |
Vis M L, Sheath R G. 1997. Biogeography of Batrachospermum gelatinosum (Batrachospermales, Rhodophyta) in North America based on molecular and morphological data. Journal of Phycology, 33(3): 520-526.
DOI:10.1111/j.0022-3646.1997.00520.x |
Xie S L, Shi Z X. 2003. Thorea (Thoreaceae, Rhodophyta) in China. Acta Hydrobiologica Sinica, 27(6): 631-634.
(in Chinese with English abstract) |
Xie S L. 2001. Studies on Batrachospermales (Rhodophyta) from China. Doctoral dissertation, Institute of Hydrobiology, Chinese Academy of Sciences. (in Chinese with English abstract)
|
Zucchi M R, Necchi O Jr. 2003. Blue-greenish acrochaetioid algae in freshwater habitats are "Chantransia" stages of Batrachospermales sensu lato (Rhodophyta). Cryptogamie Algologie, 24(2): 117-131.
|
 2021, Vol. 39
2021, Vol. 39