Institute of Oceanology, Chinese Academy of Sciences
Article Information
- FANG Kunpeng, NAN Fangru, FENG Jia, Lü Junping, LIU Qi, LIU Xudong, XIE Shulian
- Virescentia guangxiensis (Batrachospermales, Rhodophyta): a new freshwater red algal species from South China
- Journal of Oceanology and Limnology, 39(4): 1538-1546
- http://dx.doi.org/10.1007/s00343-020-0225-0
Article History
- Received Jun. 8, 2020
- accepted in principle Jul. 14, 2020
- accepted for publication Sep. 18, 2020
Red algae (Rhodophyta) primarily inhabit marine environments, and freshwater red algae account for only 3% of species diversity in the division (Sheath and Vis, 2015). The Batrachospermales is an order that encompasses 70% of all freshwater red algal diversity, and Batrachospermum is the most speciesrich genus having at least 112 species (Kumano, 2002). Based on morphological research and the application of molecular technology, seven sections of Batrachospermum have been raised to the genus level (Entwisle et al., 2009; Salomaki et al., 2014; Necchi et al., 2018, 2019a, b ; Vis et al., 2020). Batrachospermales is widely distributed throughout China: 33 species have been discovered in China, of which 12 are endemic. Nonetheless, of these 33 species, molecular data are accessible for only 13, and the remaining species have only a morphological description (Xie et al., 2020).
Genus Virescentia, which belongs to the order Batrachospermales (Nemaliophycidae, Rhodophyta), was recently established as a genus ofBatrachospermum section Virescentia in 2018 based on morphological characteristics, geographic distribution, and DNA sequence data (Necchi et al., 2018). Section Virescentia was proposed by Sirodot based on a combination of characters: greenish thalli and carpogonia with cylindrical and stalked trichogynes (Sirodot, 1873). Seven new species of this section were described and illustrated in the monograph on Batrachospermum and represent a starting point for the study of this section (Sirodot, 1884). Subsequent morphometric analyses of type specimens and North American populations resulted in the proposal of many synonymies and the recognition of only two species, Batrachospermum helminthosa Bory and B. elegans Sirodot (Sheath et al., 1994). Kumano (2002) recognized eleven species within the section. In recent years, morphological characteristics (size of carpogonia, carposporophytes, and carposporangia) of samples from Brazil, North America and Europe have been shown to differ significantly, and DNA sequence data have suggested that there is considerable genetic variation in the genus and that it is partitioned geographically (Hanyuda et al., 2004; Rueness, 2010; De Castro Agostinho and Necchi, 2014). In order to resolve the phylogenetic relationships among the species of section Virescentia, the genus Virescentia was established in 2018, and the species of Virescentia were circumscribed on the basis of morphological characteristics (shape of fascicles, occurrence of secondary fascicles, disposition of carpogonial branches, and size of carpogonia) (Necchi et al., 2018).
At present, there are six taxonomically accepted species of genus Virescentia (V. crispata (Kumano et Ratnasabapathy) Necchi, Agostinho et Vis; V. gulbenkiana (Reis) Necchi, Agostinho et Vis; V. helminthosa (Bory) Necchi, Agostinho et Vis; V. viride-americana Necchi, Agostinho et Vis; V. viride-brasiliensis (Necchi et Agostinho) Necchi, Vis et Agostinho; and V. vogesiaca (Schultz ex Skuja) Necchi, Agostinho et Vis), of which V. crispata and V.gulbenkianahave been distinguished in morphology only (Necchi et al., 2018). In China, only one taxon, V. helminthosa, has been reported in morphology (Shi, 2006; Xie et al., 2020). In this paper, a new species of Virescentia from Guangxi, China, V. guangxiensis, is described and illustrated based on morphological and molecular evidence. This study expands the known species diversity and geographical distribution of freshwater Rhodophyta in China.
2 MATERIAL AND METHOD 2.1 Sample collectionAlgal specimens were collected in December 2019 from the spring water of Baimo Cave in Bama County, Guangxi, South China (24°18′2″N, 107°5′58″E, altitude 260.5 m). Spring water comes from Baimo Cave and is also the source of the Panyang River. It is located on the shaded side of the river that receives weak sunlight, and spring water gushes out all year round and flows into the Panyang River. Water temperature (WT), pH, atmospheric pressure (AP), dissolved oxygen (DO), electronic conductivity (EC), and total dissolved solids (TDS) were measured using handheld meters (YSI Professional Plus Multiparameter Water Quality Instrument 19E102487, YSI Incorporated, Brannum Lane Yellow Springs, Ohio, USA). The samples were washed with sterile water to remove impurities and stored at -80 ℃ after quick-freezing in liquid nitrogen. Specimens for DNA analysis were preserved in silica desiccant. Some voucher specimens were preserved in 4% formaldehyde for morphological analyses; the remaining materials were pressed as herbarium voucher specimens and preserved at the Herbarium of Shanxi University (SXU) (Thiers, 2020), Shanxi University, Taiyuan, Shanxi, China.
2.2 Morphological observation and sequence amplificationMorphological analyses were performed using an Olympus BX-43 microscope equipped with a chargecoupled device (DP72; Olympus, Tokyo, Japan) for photography.
For DNA extraction, tissue dried in silica gel was homogenized by grinding in liquid nitrogen with a mortar and pestle, and total DNA was extracted following the protocol of Saunders (1993) with modifications described in Vis and Sheath (1997). A 1 282-bp fragment of the plastid-encoded ribulose- 1, 5-bisphosphate carboxylase-oxygenase large subunit gene (rbcL) and a 664-bp barcode region near the 5′ end of the cox1 gene (COI-5P) were chosen for amplification in 20-μL volumes containing 15.3-μL ddH2O, 2-μL 10× buffer, 2.0-μL 2.5-mmol/L dNTPs, 0.2-μL Taq DNA polymerase (all from Sangon Biotech Co., Ltd., China), 2.0 μL of each primer (10 mmol/L), and 0.5 μL of genomic DNA. The rbcL gene was polymerase chain reaction (PCR) amplified using the F160 and rbcLR primers (Vis et al., 1998) with the following cycle conditions: 95 ℃ for 2 min; 35 cycles of 93 ℃ for 1 min, 47 ℃ for 1 min, and 72 ℃ for 2 min; and a final hold for 2 min at 72 ℃. The COI-5P gene was PCR amplified using the GazF1 and GazR1 primers (Saunders, 2005) with the following cycle conditions: 94 ℃ for 2 min; 35 cycles of 94 ℃ for 30 s, 47 ℃ for 30 s, and 72 ℃ for 1 min; and a final hold for 7 min at 72 ℃.
2.3 Sequencing and data analysisThe PCR products were purified using a SanPrep column DNA gel purification kit (Sangon, China) and then submitted to BGI Tech Corporation (Beijing, China) for sequencing with both PCR primers using an ABI 3730XL sequencer. Contigs were assembled and edited using Sequencher 4.10.1 and submitted to GenBank.
For phylogenetic analyses, sequence data for order Batrachospermales were downloaded from GenBank (Supplementary Table S1), and Thorea hispida was used as the outgroup (Vis and Entwisle, 2000; Entwisle et al., 2009; Chapuis et al., 2017). Sequences were aligned in ClustalX 2.0 (Thompson et al., 1997), and pairwise distances between specimens of Batrachospermaceae were computed in MEGA 5.0 (Tamura et al., 2011). The aligned sequences were used to construct phylogenetic trees. Appropriate models of sequence evolution for our datasets were determined with a hierarchical likelihood ratio test performed in Modeltest version 3.7 (Posada and Buckley, 2004). The parameters for the rbcL maximum likelihood (ML) analyses were as follows: GTR+I+G model; gamma distribution=0.666 3; proportion of invariable sites=0.475 9; base frequencies A=0.385 4, C=0.105 6, G=0.136 3, and T=0.372 7; and rate matrix A–C=3.506 1, A–G=5.628 0, A– T=0.796 7, C–G=3.047 8, and C–T=18.856 4. The parameters for the COI-5P ML analyses were as follows: GTR+I+G model; gamma distribution= 0.471 3; proportion of invariable sites=0.411 1; base frequencies A=0.334 3, C=0.093 0, G=0.098 1, and T=0.474 6; and rate matrix A–C=1.600 0, A– G=9.313 4, A–T=0.144 5, C–G=3.795 6, and C– T=20.916 5. Phylogenetic inferences were made using the neighbor-joining method (NJ) in MEGA 5.0 (Tamura et al., 2011) and Bayesian inference (BI) in MrBayes version 3.2 (Ronquist and Huelsenbeck, 2003), and ML trees were constructed using PHYML (Felsenstein, 1981; Guindon and Gascuel, 2003). Both BI and ML analyses were performed using a general time reversible model with a gamma distribution. The BI analysis was run for 5 000 000 generations until the standard deviation of the split frequencies was less than 0.01. The ML analysis was performed using the rapid bootstrapping and best tree search algorithm for 1 000 repetitions. The resulting phylogenetic trees were edited using Figtree V.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
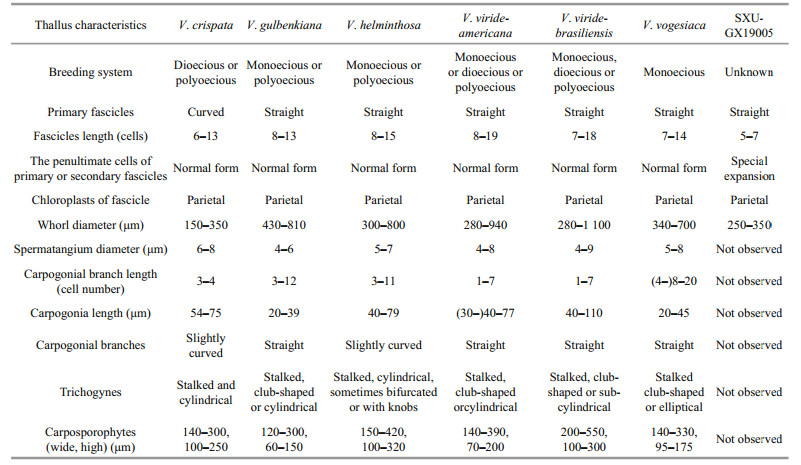
3 RESULT 3.1 Morphological observationVegetative structures were identified and illustrated (Fig. 1) based on the morphological and morphometric characters described in previous literature (Sirodot, 1884; Kumano, 2002; De Castro Agostinho and Necchi, 2014; Necchi et al., 2018), and a morphological comparison of the present sample with six previously reported species of Virescentia is provided (Table 1). Because neither spermatangia nor carpogonia were observed, distinctions in these structures between the present specimen and others were unclear. The specimen broadly overlapped previously described species for most morphometric and morphological characters: greenish thalli, irregular branching, welldeveloped whorls, few and sparse secondary fascicles, and a brownish main axis. The specimen was distinguished based on some uncertain morphological characters, including the presence of special expansion cells with a variable shape, obovoid (15–25 μm in diameter, 27–34-μm long), spherical (18–25 μm in diameter), or pear-shaped (15–20 μm in diameter, 20–30-μm long), located in the penultimate cells of primary or secondary fascicles, or rarely terminal on primary fascicles, which were not observed in other species of Virescentia. In addition, its small whorls (250–350-μm wide) and short primary fascicles (5–7 cell stories, branched 2–3 times) were also different.

|
| Fig.1 Morphological structures of V. guangxiensis a. the habit of whole plants; b. a main axis with spherical-shaped whorls; c. cortical filament cells (arrowheads); d. a node showing brown pigmented axial cell (arrowheads); e. sparsely secondary fascicles (arrowheads), dichotomously branched; f. obovoid cells (arrowheads) terminal on primary fascicles; g. cells of fascicle with chloroplasts, parietal, often dissected into spirally arranged ribbons (white arrow); h–i. cells with special morphological characteristics (arrowheads): special expansion, obovoid, spherical, pear-shaped, locating in the penultimate cells of primary or secondary fascicles. Scale bars: 1 cm in a, 100 μm in b, and 20 μm in the rest. |
|
Sequence data for species in the order Batrachospermales were collected (49 rbcL representing 40 taxa from 15 genera and 32 COI-5P representing 29 taxa from 11 genera). The rbcL alignment in this study was 1 210 bp in length with 505 variable sites (among which 436 were parsiminformative). For COI-5P, the alignment was 664 bp in length with 331 variable sites (among which 285 were parsim-informative).
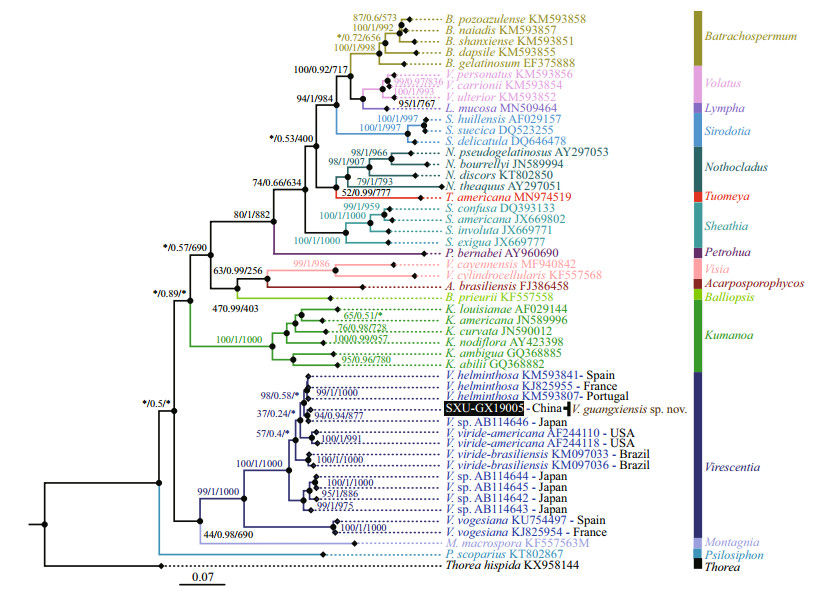
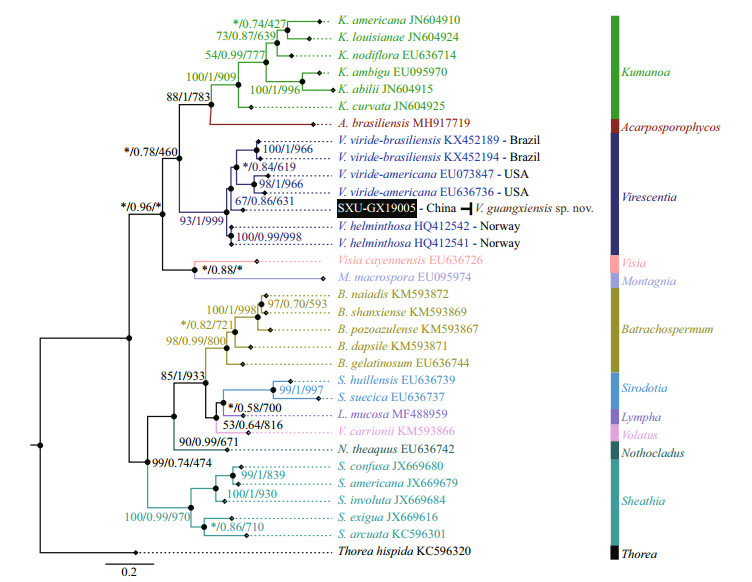
Separate phylogenetic trees based on the rbcL and COI-5P sequences were constructed using three methods (NJ, BI, and ML). All showed similar topologies, differing only in the Virescentia clade (nodes with "*") for rbcL in the ML tree and COI-5P in the NJ tree with low support values. The specimen SXU-GX19005 collected in this study was clearly placed within the Virescentia clade. In the rbcL phylogeny (Fig. 2), this specimen was placed in a well-supported clade (94/0.94/877) with Virescentia sp. (AB114646) from Japan and was sister to V. helminthosa (from Spain, France, and Portugal) with a distance of 0.77% (9.3 bp). Distance among the three V. helminthosa samples was 0.55% (6.7 bp), and that between SXU-GX19005 and Virescentia sp. AB114646 was 0.33% (4 bp). Similarly, in the COI- 5P phylogeny (Fig. 3), SXU-GX19005 was separated from other species in the Virescentia clade (67/0.86/631) with a distance of 4.97% (33 bp) from V. helminthosa, 7.38% (49 bp) from V. viridebrasiliensis, and 8.96% (59.5 bp) from V. virideamericana. These results suggested that SXUGX19005 is a new species, named Virescentia guangxiensis.
|
| Fig.2 Phylogenetic tree (BI) based on the rbc L gene sequences showing the relationships of the specimen from this study (black box) and other Batrachospermales species Support values for all analyses are shown as follows: NJ distance bootstrap/Bayesian posterior probabilities/ML bootstrap. '*' denotes the branch differed in the NJ or ML topology. |
|
| Fig.3 Phylogenetic tree (BI) based on the COI-5P gene sequences showing the relationships of the specimen from this study (black box) and other Batrachospermales species Support values for all analyses are shown as follows: NJ distance bootstrap/Bayesian posterior probabilities/ML bootstrap. '*' denotes the branch differed in the NJ or ML topology. |
Virescentia guangxiensis FANG, NAN, et XIE sp. nov. (Fig. 1a–i)
Description: plants moderately mucilaginous, greenish in color, 2–3-cm high. Whorls contiguous or separated, barrel-shaped or spherical, 250–350-μm wide, internodes 140–230 μm, main axis sometimes brownish colored, axial cells 20–40 μm in diameter, cortical filament cells cylindrical, 6–10 μm in diameter, 30 μm or more long. Primary fascicles straight, 5–7 cell stories, dichotomously branched, 2–3 times branched. Proximal cells cylindrical or barrel-shaped, 6–10 μm in diameter, 30–40-μm long, and distal cells ellipsoidal or obovoid, 5–8 μm in diameter with chloroplasts, parietal, often dissected into spirally arranged ribbons. Secondary fascicles few and sparse, unbranched or dichotomously branched, shorter than or as long as primary fascicles. Cells with special morphological characteristics, special expansion and variable in shape, obovoid (15– 25 μm in diameter, 27–34-μm long), spherical (18–25 in diameter), pear-shaped (15–20 μm in diameter, 20–30-μm long), located in the penultimate cells of primary or secondary fascicles, rarely terminal on primary fascicles. Neither spermatangia nor carpogonia were observed.
Diagnosis: the new species differs from other species in that it has special expansion cells located in the penultimate cells of primary or secondary fascicles. Diagnostic DNA sequence: rbcL and COI- 5P (accession number: MT533618 for rbcL and MT533619 for COI-5P).
Type locality: China—Guangxi, Bama County (24°18′2″N, 107°5′58″E, ASL: 260.5 m): the spring water comes from Baimo Cave and is also the source of the Panyang River (WT=21.0 ℃, pH=7.79, AP=9.086×104Pa, DO=6.66 mg/L, EC=310.1 μS/cm, TDS=218.4 mg/L).
Holotype here designated: deposited in Herbarium of Shanxi University (SXU), Shanxi University, Taiyuan, Shanxi Province, China. SXU-GX19005, Dried material prepared from reference strain GX15, Shanxi University Herbarium (SXU), Shanxi University, Taiyuan, Shanxi Province, China, December 2019 by Kunpeng FANG.
Etymology: the species epithet refers to the type locality (Guangxi, China).
4 DISCUSSIONPhylogenies based on the rbcL and COI-5P sequences revealed the genus Virescentia to be monophyletic. Within Virescentia, species clades corresponded to geographic regions, and each clade represented a species as follows: V. helminthosa (Europe), V. vogesiaca (Europe), V. viride-americana (North America), V. viride-brasiliensis (South America) and V. guangxiensis (Asia). This result was consistent with all previous molecular systematic studies (Vis et al., 1998; Hanyuda et al., 2004; Entwisle et al., 2009; De Castro Agostinho and Necchi, 2014, Necchi et al., 2018). Members of the genus can be recognized by a combination of characters: the presence of well-developed whorls; carpogonial branches well differentiated from the fascicles, straight, rarely curved; carpogonia with cylindrical, club-shaped or elliptical, stalked trichogynes; and axial, large, and dense carposporophytes (Necchi et al., 2018). Among the six taxonomically accepted species, V. helminthosa is characterized by slightly curved carpogonial branches and V. crispata by audouinelloid and curved primary and secondary fascicles. Both V. gulbenkiana and V. vogesiaca have short carpogonia (20–45 μm), but the secondary fascicles are few and sparse in V. gulbenkiana and abundant in V. vogesiaca. V. viride-americana and V. viride-brasiliensis are morphologically similar and distinguished only by geographic distribution and DNA sequence data (rbcL and COI-5P) (Necchi et al., 2018).
Virescentia guangxiensis, described here from China, represents previously undescribed diversity within Virescentia. Its morphological characteristics (greenish thalli, irregular branching, well-developed whorls, few and sparse secondary fascicles, and brownish main axis) are consistent with other species from genus Virescentia. Unfortunately, neither spermatangia nor carpogonia were observed, but this species can still be distinguished from others based on additional morphological characters. In particular, it exhibited special expansion cells with a variable shape, obovoid, spherical, or pear-shaped, located in the penultimate cells of primary or secondary fascicles, and rarely terminal on primary fascicles, which were not observed in other Virescentia species. In addition, small whorls (250–350-μm wide) and short primary fascicles (5–7 cell stories, branched 2–3 times) were also discriminative for this species. The Virescentia sp. specimen from Japan had only one rbcL sequence (rbcL-AB114646), which was similar to that of the specimen from this study, with a divergence of only 4 bp. Nonetheless, it was lack of voucher specimens for morphological observation (Hanyuda et al., 2004). Whether it is the same species as the specimens in this study is still uncertain. The remaining four sequences of other Virescentia sp. specimens from Japan formed an independent branch on the tree, representing a second potential species, consistent with previous studies (Necchi et al., 2018).
At present, only one Virescentia species, V. helminthosa, is recognized by morphology in China and distributed in two provinces, Chongqing and Zhejiang. This species has typical characteristics of V. helminthosa (whorls composed of straight fascicles and carpogonial branches slightly curved), but it lacks supporting molecular data (Shi, 2006; Xie et al., 2020). By contrast, V. guangxiensis was collected from the type locality Guangxi, which is far from Zhejiang and Chongqing. Furthermore, V. guangxiensis has small whorls (250–350-μm wide) and axial cells (20–40 μm in diameter), short primary fascicles and internodes (140–230 μm), and missing terminal hairs compared with V. helminthosa (whorls 290–440-μm wide, axial cells 60–80 μm, 6–12 primary fascicles, and 330–90-μm internodes). Thus, V. guangxiensis is different from V. helminthosa and is the second species of genus Virescentia reported in China.
Guangxi Province is located in south China and exhibits a subtropical and tropical monsoon climate with an annual average temperature of 17.5–23.5 ℃. Karst landforms are widespread in Guangxi, accounting for 37.8% of the total land area, and the groundwater is rich in calcium bicarbonate and magnesium bicarbonate. Only two taxa of order Acrochaetiales have been reported in Guangxi for Subclass Nemaliophycidae (Shi, 2006). Therefore, Virescentia guangxiensis is the first species of order Batrachospermales reported in Guangxi. The type locality Baimo Cave is located in Bama County, Hechi City. Bama County is a famous "Changshouzhixiang" (hometown of longevity) in China. "Bai Mo" means spring water outlet in the Zhuang language, and the spring in Baimo Cave is the source of the Panyanghe River. The rare morphological characteristic (special expansion cells) of the new species may be a result of environmental adaptation. However, there are no reports of similar characters in other freshwater red algae, and further research is required to determine the cause of this morphological character.
5 CONCLUSIONMorphological comparison and molecular analysis both supported the proposal of a new freshwater red algal species Virescentia guangxiensis. It is characterized morphologically by the presence of special expansion cells with a variable shape, obovoid, spherical, or pear-shaped, located in the penultimate cells of primary or secondary fascicles, rarely terminal on primary fascicles, as well as by small whorls and short primary fascicles. Phylogenetic analysis of sequence data from the rbcL and COI-5P loci supported the separation of V. guangxiensis from other species of genus Virescentia, justifying the proposal of the new species. V. guangxiensis is the first species of order Batrachospermales reported in Guangxi and the second species of genus Virescentia reported in China. The description of this new species expands the known species diversity and geographical distribution of freshwater Rhodophyta in China.
6 DATA AVAILABILITY STATEMENTThe datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENT: Dr. E. W. Christina (The Pennsylvania State University) is acknowledged for the English editing.
Chapuis I S, Necchi O Jr, Zuccarello G C, Xie S L, Aboal M, Castillo P M S, Vis M L. 2017. A new genus, Volatus and four new species of Batrachospermum sensu stricto (Batrachospermales, Rhodophyta). Phycologia, 56(4): 454-468.
DOI:10.2216/16-73.1 |
De Castro Agostinho D, Necchi O Jr. 2014. Systematics of the section Virescentia of the genus Batrachospermum (Batrachospermales, Rhodophyta) in Brazil. Phycologia, 53(6): 561-570.
DOI:10.2216/PH14-034.1 |
Entwisle T J, Vis M L, Chiasson W B, Necchi O Jr, Sherwood A R. 2009. Systematics of the Batrachospermales (Rhodophyta)—a synthesis. Journal of Phycology, 45(3): 704-715.
DOI:10.1111/j.1529-8817.2009.00686.x |
Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution, 17(6): 368-376.
DOI:10.1007/BF01734359 |
Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology, 52(5): 696-704.
DOI:10.1080/10635150390235520 |
Hanyuda T, Suzawa Y, Suzawa T, Arai S, Sato H, Ueda K, Kumano S. 2004. Biogeography and taxonomy of Batrachospermum helminthosum (Batrachospermales, Rhodophyta) in Japan inferred from rbcL gene sequences. Journal of Phycology, 40(3): 581-588.
DOI:10.1111/j.1529-8817.2004.03159.x |
Kumano S. 2002. Freshwater Red Algae of the World. Biopress Ltd, Bristol, UK. 375p.
|
Necchi O Jr, De Castro Agostinho D, Vis M L. 2018. Revision of Batrachospermum section Virescentia (Batrachospermales, Rhodophyta) with the establishment of the new genus, Virescentia stat. nov. Cryptogamie, Algologie, 39(3): 313-338.
DOI:10.7872/crya/v39.iss3.2018.313 |
Necchi O Jr, Garcia Filho A, Paiano M O. 2019a. Revision of Batrachospermum sections Acarposporophytum and Aristata (Batrachospermales, Rhodophyta) with the establishment of the new genera Acarposporophycos and Visia. Phytotaxa, 395(2): 51-65.
DOI:10.11646/phytotaxa.395.2.1 |
Necchi O Jr, Garcia Fo. A S, Paiano M O, Vis M L. 2019b. Revision of Batrachospermum section Macrospora (Batrachospermales, Rhodophyta) with the establishment of the new genus Montagnia. Phycologia, 58(6): 582-591.
DOI:10.1080/00318884.2019.1624143 |
Posada D, Buckley T R. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology, 53(5): 793-808.
DOI:10.1080/10635150490522304 |
Ronquist F, Huelsenbeck J P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12): 1 572-1 574.
DOI:10.1093/bioinformatics/btg180 |
Rueness J. 2010. DNA barcoding of select freshwater and marine red algae (Rhodophyta). Cryptogamie, Algologie, 31(4): 377-386.
|
Salomaki E D, Kwandrans J, Eloranta P, Vis M L. 2014. Molecular and morphological evidence for Sheathia gen. nov. (Batrachospermales, Rhodophyta) and three new species. Journal of Phycology, 50(3): 526-542.
DOI:10.1111/jpy.12179 |
Saunders G W. 1993. Gel purification of red algal genomic DNA: an inexpensive and rapid method for the isolation of polymerase chain reaction-friendly DNA. Journal of Phycology, 29(2): 251-254.
DOI:10.1111/j.0022-3646.1993.00251.x |
Saunders G W. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philosophical Transactions of the Royal Society B, 360(1462): 1 879-1 888.
DOI:10.1098/rstb.2005.1719 |
Sheath R G, Vis M L. 2015. Red algae. In: Freshwater Algae of North America. Academic Press, San Diego, California. p. 237–264.
|
Sheath R G, Vis M L, Cole K M. 1994. Distribution and systematics of Batrachospermum (Batrachospermales, Rhodophyta) in North America. 4. Section Virescentia. Journal of Phycology, 30(1): 108-117.
|
Shi Z X. 2006. Flora algarum sinicarum aquae dulcis, Tomus XIII, Rhodophyta, Phaeophyta. Science Press, Beijing, p. 30–77. (in Chinese)
|
Sirodot S. 1873. Nouvelle classification des algues d'eau douce du genre Batrachospermum: développement; générations alternantes. Comptes-rendus hebdomadaires des séances de l'Académie des Sciences, 76: 1 216-1 120.
|
Sirodot S. 1884. Les Batrachospermes: Organisation, Fonctions, Développement, Classification. Librairie de l'Académie de Médecine, Paris, 299p.
|
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10): 2 731-2 739.
DOI:10.1093/molbev/msr121 |
Thiers B. 2020. Index herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden's Virtual Herbarium,
|
Vis M L, Lee J, Eloranta P, Chapuis I S, Lam D W, Necchi O Jr. 2020. Paludicola gen. nov. and revision of the species formerly in Batrachospermum Section Turfosa (Batrachospermales, Rhodophyta). Journal of Phycology, 56(4): 844-861.
DOI:10.1111/jpy.13001 |
Vis M L, Saunders G W, Sheath R G, Dunse K, Entwisle T J. 1998. Phylogeny of the Batrachospermales (Rhodophyta) inferred from rbcL and 18S ribosomal DNA gene sequences. Journal of Phycology, 34(2): 341-350.
DOI:10.1046/j.1529-8817.1998.340341.x |
Vis M L, Sheath R G. 1997. Biogeography of Batrachospermum gelatinosum (Batrachospermales, Rhodophyta) in North America based on molecular and morphological data. Journal of Phycology, 33(3): 520-526.
DOI:10.1111/j.0022-3646.1997.00520.x |
Vis M L, Entwisle T J. 2000. Insights into the phylogeny of the Batrachospermales (Rhodophyta) from rbcL sequence data of Australian taxa. Journal of Phycology, 36(6): 1 175-1 182.
DOI:10.1046/j.1529-8817.2000.99224.x |
Xie S L, Qiu M Y, Nan F R, Fang K P, Han J F. 2020. Batrachospermales (Rhodophyta) of China: a catalogue and bibliography. Nova Hedwigia, 110(1-2): 37-77.
DOI:10.1127/nova_hedwigia/2020/0565 |
 2021, Vol. 39
2021, Vol. 39


